| 1 |
|
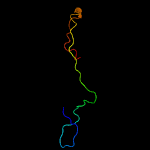
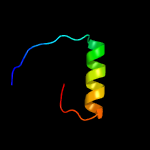
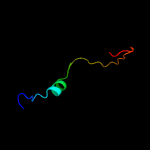
PDB 2bj7 chain A domain 1
Region: 2 - 29
Aligned: 28
Modelled: 28
Confidence: 26.1%
Identity: 18%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: CopG-like
Phyre2
| 2 |
|
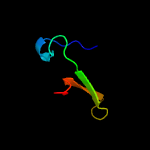
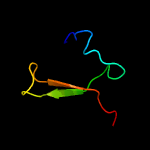
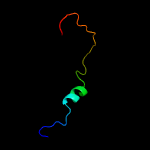
PDB 2ca9 chain B
Region: 2 - 29
Aligned: 28
Modelled: 28
Confidence: 17.3%
Identity: 21%
PDB header:transcription
Chain: B: PDB Molecule:putative nickel-responsive regulator;
PDBTitle: apo-nikr from helicobacter pylori in closed trans-2 conformation
Phyre2
| 3 |
|
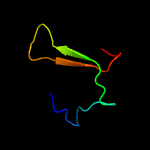
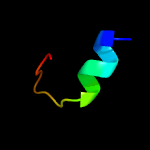
PDB 5l6m chain A
Region: 6 - 41
Aligned: 33
Modelled: 36
Confidence: 16.2%
Identity: 33%
PDB header:hydrolase
Chain: A: PDB Molecule:vapb family protein;
PDBTitle: structure of caulobacter crescentus vapbc1 (vapb1deltac:vapc1 form)
Phyre2
| 4 |
|
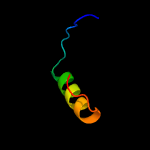
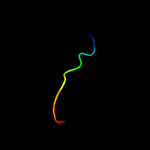
PDB 1r53 chain A
Region: 77 - 88
Aligned: 12
Modelled: 12
Confidence: 15.3%
Identity: 42%
Fold: Chorismate synthase, AroC
Superfamily: Chorismate synthase, AroC
Family: Chorismate synthase, AroC
Phyre2
| 5 |
|
PDB 6nkl chain D
Region: 6 - 26
Aligned: 20
Modelled: 21
Confidence: 13.4%
Identity: 35%
PDB header:antitoxin
Chain: D: PDB Molecule:antitoxin vapb1;
PDBTitle: 2.2 a resolution structure of vapbc-1 from nontypeable haemophilus2 influenzae
Phyre2
| 6 |
|
PDB 1v8c chain A domain 2
Region: 8 - 33
Aligned: 26
Modelled: 26
Confidence: 13.2%
Identity: 46%
Fold: TBP-like
Superfamily: MoaD-related protein, C-terminal domain
Family: MoaD-related protein, C-terminal domain
Phyre2
| 7 |
|
PDB 4lj2 chain A
Region: 35 - 88
Aligned: 48
Modelled: 54
Confidence: 12.7%
Identity: 29%
PDB header:lyase
Chain: A: PDB Molecule:chorismate synthase;
PDBTitle: crystal structure of chorismate synthase from acinetobacter baumannii2 at 3.15a resolution
Phyre2
| 8 |
|
PDB 5l6l chain D
Region: 6 - 41
Aligned: 33
Modelled: 36
Confidence: 12.7%
Identity: 36%
PDB header:hydrolase
Chain: D: PDB Molecule:vapb family protein;
PDBTitle: structure of caulobacter crescentus vapbc1 bound to operator dna
Phyre2
| 9 |
|
PDB 3tnd chain F
Region: 6 - 41
Aligned: 33
Modelled: 36
Confidence: 12.2%
Identity: 24%
PDB header:translation, toxin
Chain: F: PDB Molecule:antitoxin vapb;
PDBTitle: crystal structure of shigella flexneri vapbc toxin-antitoxin complex
Phyre2
| 10 |
|
PDB 1q5v chain B
Region: 2 - 29
Aligned: 28
Modelled: 28
Confidence: 10.8%
Identity: 21%
PDB header:transcription
Chain: B: PDB Molecule:nickel responsive regulator;
PDBTitle: apo-nikr
Phyre2
| 11 |
|
PDB 2bj3 chain D
Region: 2 - 29
Aligned: 28
Modelled: 28
Confidence: 10.3%
Identity: 18%
PDB header:transcription
Chain: D: PDB Molecule:nickel responsive regulator;
PDBTitle: nikr-apo
Phyre2
| 12 |
|
PDB 3zvk chain G
Region: 6 - 41
Aligned: 32
Modelled: 36
Confidence: 8.8%
Identity: 44%
PDB header:antitoxin/toxin/dna
Chain: G: PDB Molecule:antitoxin of toxin-antitoxin system vapb;
PDBTitle: crystal structure of vapbc2 from rickettsia felis bound to2 a dna fragment from their promoter
Phyre2
| 13 |
|
PDB 3ms6 chain A
Region: 57 - 73
Aligned: 17
Modelled: 17
Confidence: 8.5%
Identity: 47%
PDB header:protein binding
Chain: A: PDB Molecule:hepatitis b virus x-interacting protein;
PDBTitle: crystal structure of hepatitis b x-interacting protein (hbxip)
Phyre2
| 14 |
|
PDB 1sq1 chain A
Region: 66 - 88
Aligned: 23
Modelled: 23
Confidence: 8.1%
Identity: 26%
Fold: Chorismate synthase, AroC
Superfamily: Chorismate synthase, AroC
Family: Chorismate synthase, AroC
Phyre2
| 15 |
|
PDB 1p9q chain C domain 3
Region: 3 - 12
Aligned: 10
Modelled: 10
Confidence: 8.0%
Identity: 50%
Fold: Ferredoxin-like
Superfamily: EF-G C-terminal domain-like
Family: Hypothetical protein AF0491, C-terminal domain
Phyre2
| 16 |
|
PDB 1rdu chain A
Region: 2 - 12
Aligned: 11
Modelled: 11
Confidence: 7.7%
Identity: 36%
Fold: Ribonuclease H-like motif
Superfamily: Nitrogenase accessory factor-like
Family: MTH1175-like
Phyre2
| 17 |
|
PDB 2wfb chain A
Region: 2 - 7
Aligned: 6
Modelled: 6
Confidence: 6.8%
Identity: 83%
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative uncharacterized protein orp;
PDBTitle: high resolution structure of the apo form of the orange2 protein (orp) from desulfovibrio gigas
Phyre2
| 18 |
|
PDB 5z2f chain A
Region: 1 - 7
Aligned: 7
Modelled: 7
Confidence: 6.1%
Identity: 57%
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: nadph/pda bound dihydrodipicolinate reductase from paenisporosarcina2 sp. tg-14
Phyre2
| 19 |
|
PDB 4ubp chain B
Region: 9 - 45
Aligned: 33
Modelled: 37
Confidence: 5.8%
Identity: 36%
Fold: beta-clip
Superfamily: Urease, beta-subunit
Family: Urease, beta-subunit
Phyre2
| 20 |
|
PDB 1ejx chain B
Region: 9 - 45
Aligned: 33
Modelled: 37
Confidence: 5.7%
Identity: 30%
Fold: beta-clip
Superfamily: Urease, beta-subunit
Family: Urease, beta-subunit
Phyre2