| 1 |
|
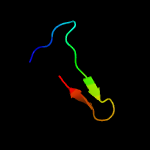
PDB 1avy chain A
Region: 163 - 200
Aligned: 34
Modelled: 38
Confidence: 45.9%
Identity: 26%
PDB header:coiled coil
Chain: A: PDB Molecule:fibritin;
PDBTitle: fibritin deletion mutant m (bacteriophage t4)
Phyre2
| 2 |
|
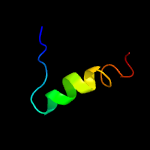
PDB 3g80 chain B
Region: 4 - 22
Aligned: 19
Modelled: 19
Confidence: 25.3%
Identity: 32%
PDB header:viral protein
Chain: B: PDB Molecule:protein b2;
PDBTitle: nodamura virus protein b2, rna-binding domain
Phyre2
| 3 |
|
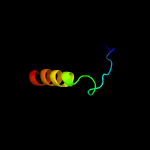
PDB 1rfo chain C
Region: 172 - 200
Aligned: 25
Modelled: 29
Confidence: 22.9%
Identity: 24%
PDB header:viral protein
Chain: C: PDB Molecule:whisker antigen control protein;
PDBTitle: trimeric foldon of the t4 phagehead fibritin
Phyre2
| 4 |
|
PDB 2lp7 chain C
Region: 172 - 204
Aligned: 29
Modelled: 33
Confidence: 17.6%
Identity: 28%
PDB header:viral protein
Chain: C: PDB Molecule:envelope glycoprotein;
PDBTitle: structure of gp41-m-mat, a membrane associated mper trimer from hiv-12 gp41.
Phyre2
| 5 |
|
PDB 1aa0 chain A
Region: 163 - 200
Aligned: 34
Modelled: 38
Confidence: 17.4%
Identity: 26%
PDB header:attachment protein
Chain: A: PDB Molecule:fibritin;
PDBTitle: fibritin deletion mutant e (bacteriophage t4)
Phyre2
| 6 |
|
PDB 1ox3 chain A
Region: 162 - 200
Aligned: 35
Modelled: 39
Confidence: 13.7%
Identity: 17%
PDB header:chaperone
Chain: A: PDB Molecule:fibritin;
PDBTitle: crystal structure of mini-fibritin
Phyre2
| 7 |
|
PDB 1k4n chain A
Region: 103 - 114
Aligned: 12
Modelled: 12
Confidence: 11.6%
Identity: 25%
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family: Hypothetical protein YecM (EC4020)
Phyre2
| 8 |
|
PDB 2knr chain A
Region: 6 - 30
Aligned: 25
Modelled: 25
Confidence: 10.4%
Identity: 12%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atc0905;
PDBTitle: solution structure of protein atu0922 from a. tumefaciens. northeast2 structural genomics consortium target att13. ontario center for3 structural proteomics target atc0905
Phyre2
| 9 |
|
PDB 5o31 chain Z
Region: 102 - 113
Aligned: 12
Modelled: 12
Confidence: 9.8%
Identity: 33%
PDB header:oxidoreductase
Chain: Z: PDB Molecule:nadh dehydrogenase [ubiquinone] 1 alpha subcomplex subunit
PDBTitle: mitochondrial complex i in the deactive state
Phyre2
| 10 |
|
PDB 6gcs chain W
Region: 102 - 112
Aligned: 11
Modelled: 11
Confidence: 9.1%
Identity: 36%
PDB header:oxidoreductase
Chain: W: PDB Molecule:nb6m subunit;
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
Phyre2
| 11 |
|
PDB 2mck chain A
Region: 192 - 199
Aligned: 8
Modelled: 8
Confidence: 8.9%
Identity: 75%
PDB header:hydrolase
Chain: A: PDB Molecule:polyprotein;
PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for murine2 norovirus cr6 ns1/2 protein
Phyre2
| 12 |
|
PDB 5lnk chain Q
Region: 102 - 112
Aligned: 11
Modelled: 11
Confidence: 7.7%
Identity: 36%
PDB header:oxidoreductase
Chain: Q: PDB Molecule:
PDBTitle: entire ovine respiratory complex i
Phyre2
| 13 |
|
PDB 1gsm chain A domain 1
Region: 191 - 217
Aligned: 27
Modelled: 27
Confidence: 7.3%
Identity: 33%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: I set domains
Phyre2
| 14 |
|
PDB 1kbl chain A domain 2
Region: 188 - 207
Aligned: 20
Modelled: 20
Confidence: 6.7%
Identity: 20%
Fold: The "swivelling" beta/beta/alpha domain
Superfamily: Phosphohistidine domain
Family: Pyruvate phosphate dikinase, central domain
Phyre2
| 15 |
|
PDB 2gqc chain A
Region: 4 - 31
Aligned: 28
Modelled: 28
Confidence: 6.6%
Identity: 21%
PDB header:hydrolase
Chain: A: PDB Molecule:rhomboid intramembrane protease;
PDBTitle: solution structure of the n-terminal domain of rhomboid intramembrane2 protease from p. aeruginosa
Phyre2
| 16 |
|
PDB 1v1c chain A
Region: 101 - 108
Aligned: 8
Modelled: 8
Confidence: 6.6%
Identity: 25%
PDB header:sh3-domain
Chain: A: PDB Molecule:obscurin;
PDBTitle: solution structure of the sh3 domain of obscurin
Phyre2
| 17 |
|
PDB 3mwz chain A
Region: 105 - 130
Aligned: 26
Modelled: 26
Confidence: 6.3%
Identity: 15%
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:sialostatin l2;
PDBTitle: crystal structure of the selenomethionine derivative of the l 22,47,2 100 m mutant of sialostatin l2
Phyre2
| 18 |
|
PDB 1r7c chain A
Region: 192 - 197
Aligned: 6
Modelled: 6
Confidence: 5.6%
Identity: 67%
PDB header:membrane protein
Chain: A: PDB Molecule:genome polyprotein;
PDBTitle: nmr structure of the membrane anchor domain (1-31) of the2 nonstructural protein 5a (ns5a) of hepatitis c virus3 (minimized average structure, sample in 50% tfe)
Phyre2
| 19 |
|
PDB 1r7g chain A
Region: 192 - 197
Aligned: 6
Modelled: 6
Confidence: 5.6%
Identity: 67%
PDB header:membrane protein
Chain: A: PDB Molecule:genome polyprotein;
PDBTitle: nmr structure of the membrane anchor domain (1-31) of the2 nonstructural protein 5a (ns5a) of hepatitis c virus3 (minimized average structure, sample in 100mm dpc)
Phyre2
| 20 |
|
PDB 4goq chain D
Region: 6 - 30
Aligned: 25
Modelled: 24
Confidence: 5.5%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a duf1491 family protein (cc_1065) from2 caulobacter crescentus cb15 at 1.87 a resolution
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|