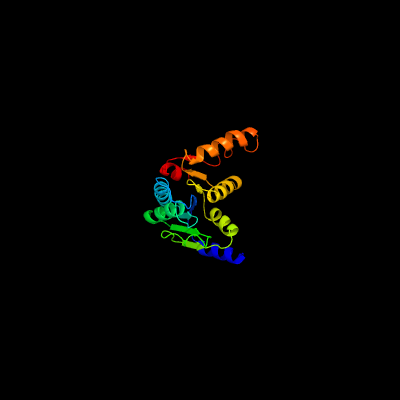
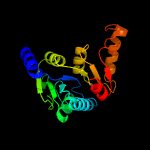
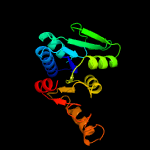
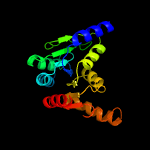
1 c5zi9B_
100.0
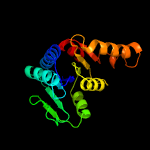
27
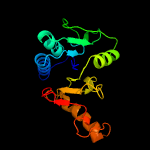
PDB header: hydrolaseChain: B: PDB Molecule: cytokinin riboside 5'-monophosphate phosphoribohydrolase;PDBTitle: crystal strcuture of type-ii log from streptomyces coelicolor a3
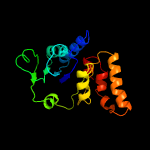
2 d1weka_
100.0
28
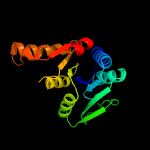
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
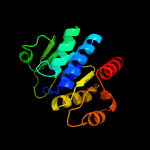
3 c3sbxC_
100.0
26
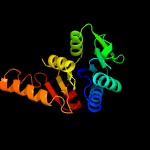
PDB header: unknown functionChain: C: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium marinum bound to adenosine 5'-monophosphate amp
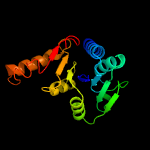
4 c5wq3A_
100.0
22
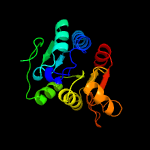
PDB header: hydrolaseChain: A: PDB Molecule: cytokinin riboside 5'-monophosphate phosphoribohydrolase;PDBTitle: crystal strcuture of type-ii log from corynebacterium glutamicum
5 c5zbjA_
100.0
23
PDB header: hydrolaseChain: A: PDB Molecule: putative cytokinin riboside 5'-monophosphatePDBTitle: crystal strcuture of type-i log from pseudomonas aeruginosa pao1
6 d1t35a_
100.0
21
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
7 c2iz6A_
100.0
34
PDB header: metal transportChain: A: PDB Molecule: molybdenum cofactor carrier protein;PDBTitle: structure of the chlamydomonas rheinhardtii moco carrier2 protein
8 d1ydha_
100.0
20
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
9 c2q4dB_
100.0
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: lysine decarboxylase-like protein at5g11950;PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at5g11950
10 c5itsD_
100.0
18
PDB header: hydrolaseChain: D: PDB Molecule: cytokinin riboside 5'-monophosphate phosphoribohydrolase;PDBTitle: crystal strcuture of log from corynebacterium glutamicum
11 c3quaA_
100.0
23
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein and possible2 molybdenum cofactor protein from mycobacterium smegmatis
12 d1rcua_
100.0
25
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
13 c1rcuB_
100.0
25
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein vt76;PDBTitle: x-ray structure of tm1055 northeast structural genomics2 consortium target vt76
14 d2q4oa1
100.0
22
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
15 c2q4oA_
100.0
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein at2g37210/t2n18.3;PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at2g37210
16 c5ajtA_
100.0
25
PDB header: hydrolaseChain: A: PDB Molecule: phosphoribohydrolase lonely guy;PDBTitle: crystal structure of ligand-free phosphoribohydrolase lonely guy from2 claviceps purpurea
17 d1weha_
100.0
25
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like
18 c3uqzB_
100.0
21
PDB header: dna binding proteinChain: B: PDB Molecule: dna processing protein dpra;PDBTitle: x-ray structure of dna processing protein a (dpra) from streptococcus2 pneumoniae
19 c3majA_
100.0
23
PDB header: dna binding proteinChain: A: PDB Molecule: dna processing chain a;PDBTitle: crystal structure of putative dna processing protein dpra from2 rhodopseudomonas palustris cga009
20 c3bq9A_
100.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted rossmann fold nucleotide-binding domain-PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
21 c6gfmA_
not modelled
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: pyrimidine/purine nucleotide 5'-monophosphate nucleosidase;PDBTitle: crystal structure of the escherichia coli nucleosidase ppnn (pppgpp-2 form)
22 c4ljkA_
not modelled
100.0
19
PDB header: dna binding proteinChain: A: PDB Molecule: dna processing chain a (dpra);PDBTitle: structural insights into the unique single-stranded dna binding mode2 of dna processing protein a from helicobacter pylori
23 c3gh1A_
not modelled
100.0
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted nucleotide-binding protein;PDBTitle: crystal structure of predicted nucleotide-binding protein from vibrio2 cholerae
24 d2nx2a1
not modelled
99.2
14
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: YpsA-like
25 c6mizC_
not modelled
98.4
19
PDB header: membrane proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: human trpm2 ion channel in an adpr-bound state
26 c6d73C_
not modelled
98.2
20
PDB header: transport proteinChain: C: PDB Molecule: transient receptor potential cation channel, subfamily m;PDBTitle: cryo-em structure of the zebrafish trpm2 channel in the presence of2 ca2+
27 c3imkA_
not modelled
98.2
18
PDB header: metal binding proteinChain: A: PDB Molecule: putative molybdenum carrier protein;PDBTitle: crystal structure of putative molybdenum carrier protein (yp_461806.1)2 from syntrophus aciditrophicus sb at 1.45 a resolution
28 c6co7C_
not modelled
98.2
20
PDB header: membrane proteinChain: C: PDB Molecule: predicted protein;PDBTitle: structure of the nvtrpm2 channel in complex with ca2+
29 c6bcoD_
not modelled
98.2
16
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of trpm4 in atp bound state with short coiled coil2 at 2.9 angstrom resolution
30 c6drkD_
not modelled
98.2
21
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel, subfamily m,PDBTitle: structure of trpm2 ion channel receptor by single particle electron2 cryo-microscopy, apo state
31 c6bcqB_
not modelled
98.1
16
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of trpm4 in atp bound state with long coiled coil at2 3.3 angstrom resolution
32 c6d73B_
not modelled
98.1
23
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel, subfamily m;PDBTitle: cryo-em structure of the zebrafish trpm2 channel in the presence of2 ca2+
33 c6nr3D_
not modelled
97.9
17
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
34 c6nr3C_
not modelled
97.9
17
PDB header: transport proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
35 c6nr3A_
not modelled
97.9
17
PDB header: transport proteinChain: A: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
36 c6nr3B_
not modelled
97.9
17
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
37 c6nr2A_
not modelled
97.7
15
PDB header: transport proteinChain: A: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with the menthol2 analog ws-12 and pi(4,5)p2
38 c6nr2C_
not modelled
97.7
15
PDB header: transport proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with the menthol2 analog ws-12 and pi(4,5)p2
39 c6nr2B_
not modelled
97.7
15
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with the menthol2 analog ws-12 and pi(4,5)p2
40 c6nr2D_
not modelled
97.7
15
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with the menthol2 analog ws-12 and pi(4,5)p2
41 c4jemA_
not modelled
96.9
16
PDB header: hydrolaseChain: A: PDB Molecule: cmp/hydroxymethyl cmp hydrolase;PDBTitle: crystal structure of milb complexed with cytidine 5'-monophosphate
42 c5vtoA_
not modelled
96.9
16
PDB header: hydrolaseChain: A: PDB Molecule: blasticidin m;PDBTitle: solution structure of blsm
43 c2khzB_
not modelled
96.7
14
PDB header: nuclear proteinChain: B: PDB Molecule: c-myc-responsive protein rcl;PDBTitle: solution structure of rcl
44 c4jenB_
not modelled
96.6
14
PDB header: hydrolaseChain: B: PDB Molecule: cmp n-glycosidase;PDBTitle: structure of clostridium botulinum cmp n-glycosidase, bcmb
45 c6evsA_
not modelled
96.5
10
PDB header: transferaseChain: A: PDB Molecule: n-deoxyribosyltransferase;PDBTitle: characterization of 2-deoxyribosyltransferase from psychrotolerant2 bacterium bacillus psychrosaccharolyticus: a suitable biocatalyst for3 the industrial synthesis of antiviral and antitumoral nucleosides
46 c6bpqA_
not modelled
96.4
15
PDB header: transport proteinChain: A: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
47 c6bpqB_
not modelled
96.4
15
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
48 c6bpqC_
not modelled
96.4
15
PDB header: transport proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
49 c6bpqD_
not modelled
96.4
15
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
50 c5nbrB_
not modelled
95.4
13
PDB header: transferaseChain: B: PDB Molecule: deoxyribosyltransferase;PDBTitle: 2-desoxiribosyltransferase from leishmania mexicana
51 d1f8ya_
not modelled
95.3
9
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase
52 d2f62a1
not modelled
95.0
13
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase
53 c3ehdA_
not modelled
93.7
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein;PDBTitle: crystal structure of conserved protein from enterococcus faecalis v583
54 c6eoaA_
not modelled
92.9
15
PDB header: flavoproteinChain: A: PDB Molecule: phosphopantothenoylcysteine decarboxylase;PDBTitle: crystal structure of hal3 from cryptococcus neoformans
55 c6hcyA_
not modelled
89.7
17
PDB header: membrane proteinChain: A: PDB Molecule: metalloreductase steap4;PDBTitle: human steap4 bound to nadp, fad, heme and fe(iii)-nta.
56 c2bonB_
not modelled
87.3
27
PDB header: transferaseChain: B: PDB Molecule: lipid kinase;PDBTitle: structure of an escherichia coli lipid kinase (yegs)
57 d1s2da_
not modelled
86.7
9
Fold: Flavodoxin-likeSuperfamily: N-(deoxy)ribosyltransferase-likeFamily: N-deoxyribosyltransferase
58 c1mvlA_
not modelled
85.5
14
PDB header: lyaseChain: A: PDB Molecule: ppc decarboxylase athal3a;PDBTitle: ppc decarboxylase mutant c175s
59 d1mvla_
not modelled
85.5
14
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
60 c5v2kA_
not modelled
83.7
10
PDB header: transferaseChain: A: PDB Molecule: udp-glycosyltransferase 74f2;PDBTitle: crystal structure of udp-glucosyltransferase, ugt74f2 (t15a), with udp2 and 2-bromobenzoic acid
61 c2ejbA_
not modelled
81.9
16
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: crystal structure of phenylacrylic acid decarboxylase from2 aquifex aeolicus
62 d1iuka_
not modelled
80.7
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
63 d1sbza_
not modelled
80.0
19
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
64 c2jzcA_
not modelled
79.6
13
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine transferase subunitPDBTitle: nmr solution structure of alg13: the sugar donor subunit of2 a yeast n-acetylglucosamine transferase. northeast3 structural genomics consortium target yg1
65 c1r0lD_
not modelled
79.1
10
PDB header: oxidoreductaseChain: D: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from zymomonas mobilis2 in complex with nadph
66 c2duwA_
not modelled
78.9
19
PDB header: ligand binding proteinChain: A: PDB Molecule: putative coa-binding protein;PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
67 d1g5qa_
not modelled
78.6
11
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
68 d2d59a1
not modelled
78.5
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
69 c6jtdB_
not modelled
78.3
16
PDB header: transferaseChain: B: PDB Molecule: c-glycosyltransferase;PDBTitle: crystal structure of tccgt1 in complex with udp
70 c6qlgD_
not modelled
76.9
18
PDB header: transferaseChain: D: PDB Molecule: flavin prenyltransferase pad1, mitochondrial;PDBTitle: crystal structure of anubix (pada1) in complex with fmn and2 dimethylallyl pyrophosphate
71 c4rheB_
not modelled
74.7
17
PDB header: lyaseChain: B: PDB Molecule: 3-octaprenyl-4-hydroxybenzoate carboxy-lyase;PDBTitle: crystal structure of ubix, an aromatic acid decarboxylase from the2 colwellia psychrerythraea 34h
72 d1p3y1_
not modelled
74.1
8
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
73 d1r0ka2
not modelled
70.7
10
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
74 c3zquA_
not modelled
70.3
13
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: structure of a probable aromatic acid decarboxylase
75 c3afoB_
not modelled
68.8
11
PDB header: transferaseChain: B: PDB Molecule: nadh kinase pos5;PDBTitle: crystal structure of yeast nadh kinase complexed with nadh
76 d2acva1
not modelled
68.6
10
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDPGT-like
77 d2jgra1
not modelled
68.6
27
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like
78 d1e3ia2
not modelled
67.5
6
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
79 c3nywD_
not modelled
66.9
22
PDB header: oxidoreductaseChain: D: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of a betaketoacyl-[acp] reductase (fabg) from2 bacteroides thetaiotaomicron
80 c3pfnB_
not modelled
66.5
13
PDB header: transferaseChain: B: PDB Molecule: nad kinase;PDBTitle: crystal structure of human nad kinase
81 c1qzuB_
not modelled
64.3
14
PDB header: lyaseChain: B: PDB Molecule: hypothetical protein mds018;PDBTitle: crystal structure of human phosphopantothenoylcysteine decarboxylase
82 d2bona1
not modelled
62.6
27
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like
83 c4yxfB_
not modelled
61.5
29
PDB header: oxidoreductaseChain: B: PDB Molecule: mups;PDBTitle: mups, a 3-oxoacyl (acp) reductase involved in mupirocin biosynthesis
84 c3wisA_
not modelled
61.4
12
PDB header: oxidoreductaseChain: A: PDB Molecule: putative dihydromethanopterin reductase (afpa);PDBTitle: crystal structure of burkholderia xenovorans dmrb in complex with fmn:2 a cubic protein cage for redox transfer
85 d1o89a2
not modelled
61.2
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
86 c3hbjA_
not modelled
60.5
12
PDB header: transferaseChain: A: PDB Molecule: flavonoid 3-o-glucosyltransferase;PDBTitle: structure of ugt78g1 complexed with udp
87 c6jlsA_
not modelled
60.5
11
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative flavoprotein decarboxylase;PDBTitle: crystal structure of fmn-dependent cysteine decarboxylases tvaf from2 thioviridamide biosynthesis
88 c3urkA_
not modelled
60.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase;PDBTitle: isph in complex with propynyl diphosphate (1061)
89 c3ke8A_
not modelled
60.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase;PDBTitle: crystal structure of isph:hmbpp-complex
90 d2p1ra1
not modelled
59.7
27
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like
91 c5nlmB_
not modelled
59.0
11
PDB header: transferaseChain: B: PDB Molecule: indoxyl udp-glucosyltransferase;PDBTitle: complex between a udp-glucosyltransferase from polygonum tinctorium2 capable of glucosylating indoxyl and indoxyl sulfate
92 c3ftpD_
not modelled
58.9
16
PDB header: oxidoreductaseChain: D: PDB Molecule: 3-oxoacyl-[acyl-carrier protein] reductase;PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase from2 burkholderia pseudomallei at 2.05 a resolution
93 c6m9uA_
not modelled
58.2
16
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, short chain dehydrogenase/reductase familyPDBTitle: structure of the apo-form of 20beta-hydroxysteroid dehydrogenase from2 bifidobacterium adolescentis strain l2-32
94 c3gafF_
not modelled
58.1
19
PDB header: oxidoreductaseChain: F: PDB Molecule: 7-alpha-hydroxysteroid dehydrogenase;PDBTitle: 2.2a crystal structure of 7-alpha-hydroxysteroid dehydrogenase from2 brucella melitensis
95 c3p19A_
not modelled
57.2
23
PDB header: oxidoreductaseChain: A: PDB Molecule: putative blue fluorescent protein;PDBTitle: improved nadph-dependent blue fluorescent protein
96 d1qora2
not modelled
56.8
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
97 c2xcuC_
not modelled
56.0
17
PDB header: transferaseChain: C: PDB Molecule: 3-deoxy-d-manno-2-octulosonic acid transferase;PDBTitle: membrane-embedded monofunctional glycosyltransferase waaa of aquifex2 aeolicus, complex with cmp
98 c4n7bA_
not modelled
55.6
24
PDB header: oxidoreductaseChain: A: PDB Molecule: lytb;PDBTitle: structure of the e-1-hydroxy-2-methyl-but-2-enyl-4-diphosphate2 reductase from plasmodium falciparum
99 d2vcha1
not modelled
55.6
8
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDPGT-like
100 c3imfA_
not modelled
54.7
22
PDB header: oxidoreductaseChain: A: PDB Molecule: short chain dehydrogenase;PDBTitle: 1.99 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor'
101 c3upyB_
not modelled
54.5
13
PDB header: oxidoreductaseChain: B: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of the brucella abortus enzyme catalyzing the first2 committed step of the methylerythritol 4-phosphate pathway.
102 d1lpfa2
not modelled
54.5
26
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
103 c3ff4A_
not modelled
54.2
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein chu_1412
104 c4kmsB_
not modelled
54.1
21
PDB header: oxidoreductaseChain: B: PDB Molecule: acetoacetyl-coa reductase;PDBTitle: crystal structure of acetoacetyl-coa reductase from rickettsia felis
105 c2vq3B_
not modelled
54.0
30
PDB header: oxidoreductaseChain: B: PDB Molecule: metalloreductase steap3;PDBTitle: crystal structure of the membrane proximal oxidoreductase2 domain of human steap3, the dominant ferric reductase of3 the erythroid transferrin cycle
106 c4zu3D_
not modelled
53.9
14
PDB header: lyaseChain: D: PDB Molecule: halohydrin epoxidase b;PDBTitle: halohydrin hydrogen-halide-lyases, hheb
107 d1h6va2
not modelled
53.8
22
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
108 c2q5cA_
not modelled
53.6
15
PDB header: transcriptionChain: A: PDB Molecule: ntrc family transcriptional regulator;PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
109 c2q62A_
not modelled
53.2
15
PDB header: flavoproteinChain: A: PDB Molecule: arsh;PDBTitle: crystal structure of arsh from sinorhizobium meliloti
110 d3grsa2
not modelled
52.5
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
111 c5iz4A_
not modelled
52.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: putative short-chain dehydrogenase/reductase;PDBTitle: crystal structure of a putative short-chain dehydrogenase/reductase2 from burkholderia xenovorans
112 d1qzua_
not modelled
51.5
11
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
113 c5b1yB_
not modelled
51.0
22
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] reductase;PDBTitle: crystal structure of nadph bound carbonyl reductase from aeropyrum2 pernix
114 d1e7wa_
not modelled
50.9
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
115 d1f8fa2
not modelled
50.6
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
116 c3a14B_
not modelled
50.5
15
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
117 c2dknA_
not modelled
50.4
29
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-alpha-hydroxysteroid dehydrogenase;PDBTitle: crystal structure of the 3-alpha-hydroxysteroid dehydrogenase from2 pseudomonas sp. b-0831 complexed with nadh
118 c3t4xA_
not modelled
50.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, short chain dehydrogenase/reductase family;PDBTitle: short chain dehydrogenase/reductase family oxidoreductase from2 bacillus anthracis str. ames ancestor
119 c3nvaB_
not modelled
50.2
25
PDB header: ligaseChain: B: PDB Molecule: ctp synthase;PDBTitle: dimeric form of ctp synthase from sulfolobus solfataricus
120 c3a28H_
not modelled
49.7
13
PDB header: oxidoreductaseChain: H: PDB Molecule: l-2.3-butanediol dehydrogenase;PDBTitle: crystal structure of l-2,3-butanediol dehydrogenase