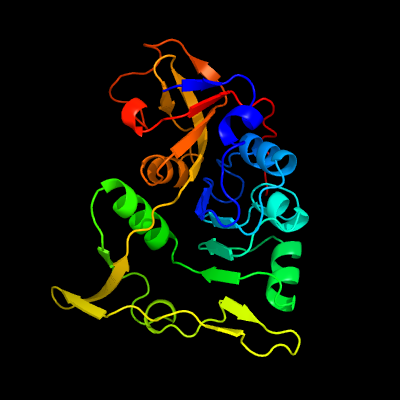
1 d1ooya2
100.0
56
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
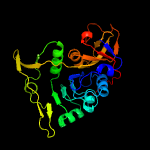
2 d2ahua2
100.0
25
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
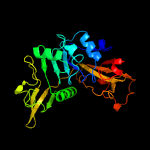
3 d1poia_
100.0
24
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
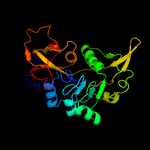
4 c3rrlC_
100.0
53
PDB header: transferaseChain: C: PDB Molecule: succinyl-coa:3-ketoacid-coenzyme a transferase subunit a;PDBTitle: complex structure of 3-oxoadipate coa-transferase subunit a and b from2 helicobacter pylori 26695
5 c6co9A_
100.0
28
PDB header: hydrolaseChain: A: PDB Molecule: probable coa-transferase alpha subunit;PDBTitle: crystal structure of rhodococcus jostii rha1 ipdab cochea-coa complex
6 c3cdkA_
100.0
61
PDB header: transferaseChain: A: PDB Molecule: succinyl-coa:3-ketoacid-coenzyme a transferasePDBTitle: crystal structure of the co-expressed succinyl-coa2 transferase a and b complex from bacillus subtilis
7 c5mzyA_
100.0
28
PDB header: lyaseChain: A: PDB Molecule: glutaconate coa-transferase family, subunit a;PDBTitle: crystal structure of the decarboxylase aiba/aibb in complex with a2 possible transition state analog
8 c4kgbB_
100.0
47
PDB header: transferaseChain: B: PDB Molecule: succinyl-coa:3-ketoacid-coenzyme a transferase;PDBTitle: structure of succinyl-coa: 3-ketoacid coa transferase from drosophila2 melanogaster
9 c1ooyA_
100.0
56
PDB header: transferaseChain: A: PDB Molecule: succinyl-coa:3-ketoacid-coenzyme a transferase,PDBTitle: succinyl-coa:3-ketoacid coa transferase from pig heart
10 c2ahvC_
100.0
25
PDB header: transferaseChain: C: PDB Molecule: putative enzyme ydif;PDBTitle: crystal structure of acyl-coa transferase from e. coli o157:h7 (ydif)-2 thioester complex with coa- 1
11 d1k6da_
100.0
43
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
12 c2hj0A_
100.0
18
PDB header: lyaseChain: A: PDB Molecule: putative citrate lyase, alfa subunit;PDBTitle: crystal structure of the putative alfa subunit of citrate lyase in2 complex with citrate from streptococcus mutans, northeast structural3 genomics target smr12 .
13 c3qlkB_
100.0
23
PDB header: transferaseChain: B: PDB Molecule: coenzyme a transferase;PDBTitle: crystal structure of ripa from yersinia pestis
14 c1xr4B_
100.0
21
PDB header: hydrolase/transferaseChain: B: PDB Molecule: putative citrate lyase alpha chain/citrate-acp transferase;PDBTitle: x-ray crystal structure of putative citrate lyase alpha chain/citrate-2 acp transferase [salmonella typhimurium]
15 c4eu4A_
100.0
23
PDB header: transferaseChain: A: PDB Molecule: succinyl-coa:acetate coenzyme a transferase;PDBTitle: succinyl-coa: acetate coa-transferase (aarch6) in complex with coa2 (hexagonal lattice)
16 c3gk7A_
100.0
17
PDB header: transferaseChain: A: PDB Molecule: 4-hydroxybutyrate coa-transferase;PDBTitle: crystal structure of 4-hydroxybutyrate coa-transferase from2 clostridium aminobutyricum
17 c2oasA_
100.0
16
PDB header: transferaseChain: A: PDB Molecule: 4-hydroxybutyrate coenzyme a transferase;PDBTitle: crystal structure of 4-hydroxybutyrate coenzyme a transferase (atoa)2 in complex with coa from shewanella oneidensis, northeast structural3 genomics target sor119.
18 c2g39A_
100.0
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: acetyl-coa hydrolase;PDBTitle: crystal structure of coenzyme a transferase from pseudomonas2 aeruginosa
19 c5vitP_
99.9
23
PDB header: transferaseChain: P: PDB Molecule: mdca;PDBTitle: crystal structure of a pseudomonas malonate decarboxylase hetero-2 tetramer in complex with malonate
20 c2nvvF_
99.9
20
PDB header: hydrolaseChain: F: PDB Molecule: acetyl-coa hydrolase/transferase family protein;PDBTitle: crystal structure of the putative acetyl-coa hydrolase/transferase2 pg1013 from porphyromonas gingivalis, northeast structural genomics3 target pgr16.
21 d1xr4a1
not modelled
99.9
22
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
22 c3eh7A_
not modelled
99.9
22
PDB header: transferaseChain: A: PDB Molecule: 4-hydroxybutyrate coa-transferase;PDBTitle: the structure of a putative 4-hydroxybutyrate coa-transferase from2 porphyromonas gingivalis w83
23 c3d3uA_
not modelled
99.9
17
PDB header: transferaseChain: A: PDB Molecule: 4-hydroxybutyrate coa-transferase;PDBTitle: crystal structure of 4-hydroxybutyrate coa-transferase (abft-2) from2 porphyromonas gingivalis. northeast structural genomics consortium3 target pgr26
24 d2g39a1
not modelled
99.9
19
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
25 c6eepA_
not modelled
98.1
26
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase from legionella2 pneumophila
26 c5uf2A_
not modelled
97.9
26
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose 5 phosphate isomerase a from neisseria2 gonorrhoeae
27 c5dbnD_
not modelled
97.8
18
PDB header: transferaseChain: D: PDB Molecule: acetate coa-transferase subunit beta;PDBTitle: crystal structure of atoda complex
28 c1m0sA_
not modelled
97.8
25
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: northeast structural genomics consortium (nesg id ir21)
29 c3l7oB_
not modelled
97.8
23
PDB header: isomeraseChain: B: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
30 d2g39a2
not modelled
97.8
20
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
31 c2f8mB_
not modelled
97.7
22
PDB header: isomeraseChain: B: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: ribose 5-phosphate isomerase from plasmodium falciparum
32 d1m0sa1
not modelled
97.6
27
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain
33 c4gmkB_
not modelled
97.6
23
PDB header: isomeraseChain: B: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose 5-phosphate isomerase from the probiotic2 bacterium lactobacillus salivarius ucc118
34 c6j1kA_
not modelled
97.5
25
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from ochrobactrum2 sp. csl1
35 c1lk5C_
not modelled
97.5
29
PDB header: isomeraseChain: C: PDB Molecule: d-ribose-5-phosphate isomerase;PDBTitle: structure of the d-ribose-5-phosphate isomerase from2 pyrococcus horikoshii
36 c1lkzB_
not modelled
97.5
27
PDB header: isomeraseChain: B: PDB Molecule: ribose 5-phosphate isomerase a;PDBTitle: crystal structure of d-ribose-5-phosphate isomerase (rpia) from2 escherichia coli.
37 c3hheA_
not modelled
97.5
18
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from bartonella2 henselae
38 c3kwmC_
not modelled
97.4
27
PDB header: isomeraseChain: C: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-isomerase a
39 d1xr4a2
not modelled
97.4
23
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
40 c5n02B_
not modelled
97.2
22
PDB header: lyaseChain: B: PDB Molecule: glutaconate coa-transferase family, subunit b;PDBTitle: crystal structure of the decarboxylase aiba/aibb c56s variant
41 c1uj6A_
not modelled
97.1
19
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: crystal structure of thermus thermophilus ribose-5-phosphate isomerase2 complexed with arabinose-5-phosphate
42 d1uj4a1
not modelled
97.0
18
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain
43 c2pjmA_
not modelled
96.7
15
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
44 c3cdkD_
not modelled
96.7
19
PDB header: transferaseChain: D: PDB Molecule: succinyl-coa:3-ketoacid-coenzyme a transferasePDBTitle: crystal structure of the co-expressed succinyl-coa2 transferase a and b complex from bacillus subtilis
45 c4nmlA_
not modelled
96.5
23
PDB header: isomeraseChain: A: PDB Molecule: ribulose 5-phosphate isomerase;PDBTitle: 2.60 angstrom resolution crystal structure of putative ribose 5-2 phosphate isomerase from toxoplasma gondii me49 in complex with dl-3 malic acid
46 c3u7jA_
not modelled
96.3
22
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
47 d1lk5a1
not modelled
96.2
28
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain
48 c4x84C_
not modelled
96.1
32
PDB header: isomeraseChain: C: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from pseudomonas2 aeruginosa
49 c6conF_
not modelled
96.0
25
PDB header: hydrolaseChain: F: PDB Molecule: coa-transferase subunit beta;PDBTitle: crystal structure of mycobacterium tuberculosis ipdab
50 d1poib_
not modelled
95.4
20
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase beta subunit-like
51 d1ooya1
not modelled
95.2
27
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase beta subunit-like
52 c1xtzA_
not modelled
95.1
24
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase;PDBTitle: crystal structure of the s. cerevisiae d-ribose-5-phosphate isomerase:2 comparison with the archeal and bacterial enzymes
53 c6co6B_
not modelled
95.1
23
PDB header: hydrolaseChain: B: PDB Molecule: probable coa-transferase beta subunit;PDBTitle: crystal structure of rhodococcus jostii rha1 ipdab
54 d2ahua1
not modelled
93.3
28
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase beta subunit-like
55 c3rrlD_
not modelled
93.2
22
PDB header: transferaseChain: D: PDB Molecule: succinyl-coa:3-ketoacid-coenzyme a transferase subunit b;PDBTitle: complex structure of 3-oxoadipate coa-transferase subunit a and b from2 helicobacter pylori 26695
56 c3ecsD_
not modelled
91.2
20
PDB header: translationChain: D: PDB Molecule: translation initiation factor eif-2b subunit alpha;PDBTitle: crystal structure of human eif2b alpha
57 d1t9ka_
not modelled
90.3
23
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like
58 c5b04B_
not modelled
89.3
25
PDB header: translationChain: B: PDB Molecule: translation initiation factor eif-2b subunit alpha;PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
59 c6qg0G_
not modelled
88.1
18
PDB header: translationChain: G: PDB Molecule: translation initiation factor eif-2b subunit delta;PDBTitle: structure of eif2b-eif2 (phosphorylated at ser51) complex (model 1)
60 d1t5oa_
not modelled
85.9
11
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like
61 c5b04G_
not modelled
85.3
17
PDB header: translationChain: G: PDB Molecule: probable translation initiation factor eif-2b subunitPDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
62 c4zemB_
not modelled
81.3
16
PDB header: translationChain: B: PDB Molecule: translation initiation factor eif2b-like protein,PDBTitle: crystal structure of eif2b beta from chaetomium thermophilum
63 d1o8bb1
not modelled
79.1
26
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain
64 c6i7tB_
not modelled
78.5
20
PDB header: translationChain: B: PDB Molecule: translation initiation factor eif-2b subunit alpha;PDBTitle: eif2b:eif2 complex
65 c6i3mD_
not modelled
77.4
17
PDB header: translationChain: D: PDB Molecule: translation initiation factor eif-2b subunit delta;PDBTitle: eif2b:eif2 complex, phosphorylated on eif2 alpha serine 52.
66 c5b04C_
not modelled
74.0
10
PDB header: translationChain: C: PDB Molecule: probable translation initiation factor eif-2b subunit beta;PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
67 c3a11D_
not modelled
72.9
17
PDB header: isomeraseChain: D: PDB Molecule: translation initiation factor eif-2b, delta subunit;PDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
68 d1vb5a_
not modelled
68.6
22
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like
69 d2a0ua1
not modelled
65.3
12
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like
70 c4zeoH_
not modelled
64.7
22
PDB header: translationChain: H: PDB Molecule: translation initiation factor eif-2b-like protein;PDBTitle: crystal structure of eif2b delta from chaetomium thermophilum
71 c6a34B_
not modelled
64.5
15
PDB header: isomeraseChain: B: PDB Molecule: putative methylthioribose-1-phosphate isomerase;PDBTitle: crystal structure of 5-methylthioribose 1-phosphate isomerase from2 pyrococcus horikoshii ot3 - form i
72 c4oqqA_
not modelled
62.1
17
PDB header: transcriptionChain: A: PDB Molecule: deoxyribonucleoside regulator;PDBTitle: structure of the effector-binding domain of deoxyribonucleoside2 regulator deor from bacillus subtilis
73 c6ezoD_
not modelled
56.0
18
PDB header: membrane proteinChain: D: PDB Molecule: translation initiation factor eif-2b subunit beta;PDBTitle: eukaryotic initiation factor eif2b in complex with isrib
74 c2yvkA_
not modelled
55.4
15
PDB header: isomeraseChain: A: PDB Molecule: methylthioribose-1-phosphate isomerase;PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
75 c3wrwE_
not modelled
53.1
17
PDB header: transferaseChain: E: PDB Molecule: tm-1 protein;PDBTitle: crystal structure of the n-terminal domain of resistance protein
76 c3smaD_
not modelled
52.7
25
PDB header: transferaseChain: D: PDB Molecule: frbf;PDBTitle: a new n-acetyltransferase fold in the structure and mechanism of the2 phosphonate biosynthetic enzyme frbf
77 c6mb6A_
not modelled
48.8
31
PDB header: transferaseChain: A: PDB Molecule: aac(3)-iiib protein;PDBTitle: aac-iiib binary with coash
78 c5ht0B_
not modelled
47.1
22
PDB header: transferaseChain: B: PDB Molecule: aminoglycoside acetyltransferase hmb0005;PDBTitle: crystal structure of an antibiotic_nat family aminoglycoside2 acetyltransferase hmb0038 from an uncultured soil metagenomic sample3 in complex with coenzyme a
79 c3nzeB_
not modelled
45.2
15
PDB header: transcription regulatorChain: B: PDB Molecule: putative transcriptional regulator, sugar-binding family;PDBTitle: the crystal structure of a domain of a possible sugar-binding2 transcriptional regulator from arthrobacter aurescens tc1.
80 d2nyga1
not modelled
43.7
23
Fold: TTHA0583/YokD-likeSuperfamily: TTHA0583/YokD-likeFamily: Aminoglycoside 3-N-acetyltransferase-like
81 d2r5fa1
not modelled
43.6
10
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: SorC sugar-binding domain-like
82 c6i3mF_
not modelled
42.7
11
PDB header: translationChain: F: PDB Molecule: translation initiation factor eif-2b subunit beta;PDBTitle: eif2b:eif2 complex, phosphorylated on eif2 alpha serine 52.
83 c5dboA_
not modelled
42.0
26
PDB header: translationChain: A: PDB Molecule: translation initiation factor eif-2b-like protein;PDBTitle: crystal structure of the tetrameric eif2b-beta2-delta2 complex from c.2 thermophilum
84 c4r9nA_
not modelled
42.0
16
PDB header: transcriptionChain: A: PDB Molecule: lmo0547 protein;PDBTitle: deor family transcriptional regulator from listeria monocytogenes.
85 d2gnpa1
not modelled
41.9
11
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: SorC sugar-binding domain-like
86 d3efba1
not modelled
31.3
21
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: SorC sugar-binding domain-like
87 c3e4fB_
not modelled
30.3
17
PDB header: transferaseChain: B: PDB Molecule: aminoglycoside n3-acetyltransferase;PDBTitle: crystal structure of ba2930- a putative aminoglycoside n3-2 acetyltransferase from bacillus anthracis
88 c1dbgA_
not modelled
28.3
16
PDB header: lyaseChain: A: PDB Molecule: chondroitinase b;PDBTitle: crystal structure of chondroitinase b
89 d1o8ba1
not modelled
25.0
14
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain
90 c2w48D_
not modelled
24.6
15
PDB header: transcriptionChain: D: PDB Molecule: sorbitol operon regulator;PDBTitle: crystal structure of the full-length sorbitol operon2 regulator sorc from klebsiella pneumoniae
91 d2grea1
not modelled
23.6
26
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Aminopeptidase/glucanase lid domainFamily: Aminopeptidase/glucanase lid domain
92 c5mvrA_
not modelled
23.6
18
PDB header: transferaseChain: A: PDB Molecule: trna threonylcarbamoyladenosine biosynthesis protein tsae;PDBTitle: crystal structure of bacillus subtilus ydib
93 c6bc3A_
not modelled
22.2
15
PDB header: transferase/antibioticChain: A: PDB Molecule: aac 3-vi protein;PDBTitle: cryo x-ray structure of sisomicin bound aac-via
94 c3izbP_
not modelled
21.2
13
PDB header: ribosomeChain: P: PDB Molecule: 40s ribosomal protein rps11 (s17p);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
95 c6mn5A_
not modelled
20.4
25
PDB header: transferase/antibioticChain: A: PDB Molecule: aminoglycoside n(3)-acetyltransferase, aac(3)-iva;PDBTitle: crystal structure of aminoglycoside acetyltransferase aac(3)-iva,2 h154a mutant, in complex with gentamicin c1a
96 c3iz6P_
not modelled
20.2
19
PDB header: ribosomeChain: P: PDB Molecule: 40s ribosomal protein s11 (s17p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
97 d1ofla_
not modelled
19.9
16
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Chondroitinase B
98 d1jb7a3
not modelled
18.6
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB
99 d1soua_
not modelled
18.5
10
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: Methenyltetrahydrofolate synthetase