1 c4pmnB_
100.0
99
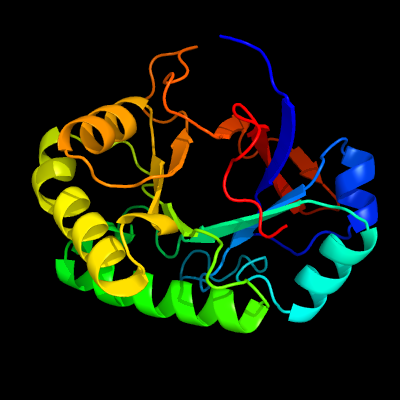
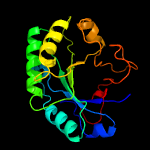
PDB header: unknown functionChain: B: PDB Molecule: tat-secreted protein rv2525c;PDBTitle: crystal structure of the mycobacterium tuberculosis tat-secreted2 protein rv2525c in complex with mes (monoclinic crystal form i)
2 c1sfsA_
100.0
21
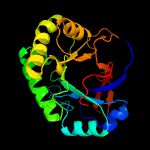
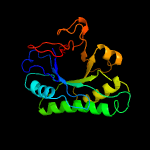
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: 1.07 a crystal structure of an uncharacterized b. stearothermophilus2 protein
3 d1sfsa_
100.0
21
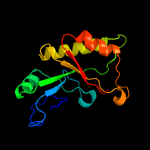
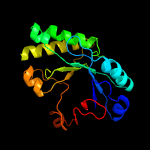
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: 1,4-beta-N-acetylmuraminidase
4 d1jfxa_
97.8
21
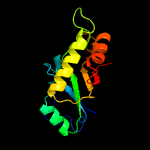
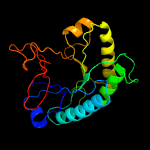
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: 1,4-beta-N-acetylmuraminidase
5 c2nw0B_
97.7
16
PDB header: hydrolaseChain: B: PDB Molecule: plyb;PDBTitle: crystal structure of a lysin
6 c4krtA_
97.7
15
PDB header: hydrolaseChain: A: PDB Molecule: autolytic lysozyme;PDBTitle: x-ray structure of endolysin from clostridium perfringens phage2 phism101
7 c2wagA_
97.5
10
PDB header: hydrolaseChain: A: PDB Molecule: lysozyme, putative;PDBTitle: the structure of a family 25 glycosyl hydrolase from2 bacillus anthracis.
8 c5a6sA_
97.5
17
PDB header: structural proteinChain: A: PDB Molecule: endolysin;PDBTitle: crystal structure of the ctp1l endolysin reveals how its2 activity is regulated by a secondary translation product
9 c4jz5A_
97.3
17
PDB header: hydrolaseChain: A: PDB Molecule: gp26;PDBTitle: high-resolution structure of catalytic domain of endolysin ply40 from2 bacteriophage p40 of listeria monocytogenes
10 c2x8rE_
97.2
18
PDB header: hydrolaseChain: E: PDB Molecule: glycosyl hydrolase;PDBTitle: the structure of a family gh25 lysozyme from aspergillus2 fumigatus
11 c4ff5A_
96.7
18
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase 25;PDBTitle: structure basis of a novel virulence factor ghip a glycosyl hydrolase2 25 of streptococcus pneumoniae participating in host cell invasion
12 c2wwcA_
96.4
16
PDB header: hydrolaseChain: A: PDB Molecule: 1,4-beta-n-acetylmuramidase;PDBTitle: 3d-structure of the modular autolysin lytc from2 streptococcus pneumoniae in complex with synthetic3 peptidoglycan ligand
13 c5jipB_
95.2
15
PDB header: hydrolaseChain: B: PDB Molecule: cortical-lytic enzyme;PDBTitle: crystal structure of the clostridium perfringens spore cortex lytic2 enzyme slem
14 d2j8ga2
86.7
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: 1,4-beta-N-acetylmuraminidase
15 c2j8fA_
72.5
11
PDB header: hydrolaseChain: A: PDB Molecule: lysozyme;PDBTitle: crystal structure of the modular cpl-1 endolysin complexed with a2 peptidoglycan analogue (e94q mutant in complex with a disaccharide-3 pentapeptide)
16 c2pq4B_
57.7
32
PDB header: chaperone/oxidoreductaseChain: B: PDB Molecule: periplasmic nitrate reductase precursor;PDBTitle: nmr solution structure of napd in complex with napa1-352 signal peptide
17 d1i60a_
56.7
11
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like
18 c5tnvA_
48.7
17
PDB header: isomeraseChain: A: PDB Molecule: ap endonuclease, family protein 2;PDBTitle: crystal structure of a xylose isomerase-like tim barrel protein from2 mycobacterium smegmatis in complex with magnesium
19 c1p84E_
47.5
18
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
20 c2fynO_
46.3
36
PDB header: oxidoreductaseChain: O: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
21 c3dx5A_
not modelled
43.7
11
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein asbf;PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
22 d1bxca_
not modelled
42.3
16
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
23 d1k77a_
not modelled
42.1
13
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Hypothetical protein YgbM (EC1530)
24 c2e76D_
not modelled
41.3
12
PDB header: photosynthesisChain: D: PDB Molecule: cytochrome b6-f complex iron-sulfur subunit;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
25 c3s6dA_
not modelled
37.0
16
PDB header: isomeraseChain: A: PDB Molecule: putative triosephosphate isomerase;PDBTitle: crystal structure of a putative triosephosphate isomerase from2 coccidioides immitis
26 d2q02a1
not modelled
37.0
7
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like
27 d2j85a1
not modelled
36.9
21
Fold: STIV B116-likeSuperfamily: STIV B116-likeFamily: STIV B116-like
28 c6cwhB_
not modelled
35.9
21
PDB header: sugar binding proteinChain: B: PDB Molecule: surface (s-) layer glycoprotein;PDBTitle: crystal structure of spaa-slh in complex with 4,6-pyr-beta-d-mannacome2 (p1)
29 c2fyuE_
not modelled
33.2
18
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit,PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
30 c1vioA_
not modelled
32.7
20
PDB header: lyaseChain: A: PDB Molecule: ribosomal small subunit pseudouridine synthase a;PDBTitle: crystal structure of pseudouridylate synthase
31 c3ju2A_
not modelled
31.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein smc04130;PDBTitle: crystal structure of protein smc04130 from sinorhizobium meliloti 1021
32 d1tz9a_
not modelled
31.3
17
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: UxuA-like
33 c6fo2R_
not modelled
29.9
16
PDB header: membrane proteinChain: R: PDB Molecule: cytochrome b-c1 complex subunit rieske, mitochondrial;PDBTitle: cryoem structure of bovine cytochrome bc1 with no ligand bound
34 c3ogrA_
not modelled
28.2
23
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: complex structure of beta-galactosidase from trichoderma reesei with2 galactose
35 c2qw5B_
not modelled
25.9
15
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase-like tim barrel;PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
36 d2g0wa1
not modelled
25.4
10
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like
37 c3c85D_
not modelled
25.4
15
PDB header: transport proteinChain: D: PDB Molecule: putative glutathione-regulated potassium-efflux systemPDBTitle: crystal structure of trka domain of putative glutathione-regulated2 potassium-efflux kefb from vibrio parahaemolyticus
38 c1xc6A_
not modelled
25.2
22
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: native structure of beta-galactosidase from penicillium sp. in complex2 with galactose
39 d1ltqa1
not modelled
25.1
17
Fold: HAD-likeSuperfamily: HAD-likeFamily: phosphatase domain of polynucleotide kinase
40 d1bxba_
not modelled
23.7
16
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
41 c5ak1A_
not modelled
23.1
15
PDB header: hydrolaseChain: A: PDB Molecule: carbohydrate binding family 6;PDBTitle: the penta-modular cellulosomal arabinoxylanase ctxyl5a2 structure as revealed by x-ray crystallography
42 c3cqkB_
not modelled
23.1
8
PDB header: isomeraseChain: B: PDB Molecule: l-ribulose-5-phosphate 3-epimerase ulae;PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
43 c6daoB_
not modelled
21.5
17
PDB header: lyaseChain: B: PDB Molecule: trans-o-hydroxybenzylidenepyruvate hydratase-aldolase;PDBTitle: nahe wt selenomethionine
44 c4wcxC_
not modelled
20.5
11
PDB header: lyaseChain: C: PDB Molecule: biotin and thiamin synthesis associated;PDBTitle: crystal structure of hydg: a maturase of the [fefe]-hydrogenase
45 d1xima_
not modelled
19.8
17
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
46 c2ogfD_
not modelled
19.0
19
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein mj0408;PDBTitle: crystal structure of protein mj0408 from methanococcus jannaschii,2 pfam duf372
47 c4kqxB_
not modelled
18.8
15
PDB header: oxidoreductase/oxidoreductase inhibitorChain: B: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: mutant slackia exigua kari ddv in complex with nad and an inhibitor
48 c5jp6A_
not modelled
18.6
23
PDB header: hydrolaseChain: A: PDB Molecule: putative polysaccharide deacetylase;PDBTitle: bdellovibrio bacteriovorus peptidoglycan deacetylase bd3279
49 d1qt1a_
not modelled
18.5
19
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
50 d1xlma_
not modelled
17.3
16
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
51 c3t7vA_
not modelled
16.9
8
PDB header: transferaseChain: A: PDB Molecule: methylornithine synthase pylb;PDBTitle: crystal structure of methylornithine synthase (pylb)
52 c5hmqE_
not modelled
14.9
13
PDB header: lyaseChain: E: PDB Molecule: 4-hydroxyphenylpyruvate dioxygenase;PDBTitle: xylose isomerase-like tim barrel/4-hydroxyphenylpyruvate dioxygenase2 fusion protein
53 d2obpa1
not modelled
14.3
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ReutB4095-like
54 c2fhoA_
not modelled
14.2
18
PDB header: rna binding proteinChain: A: PDB Molecule: spliceosomal protein sf3b155;PDBTitle: nmr solution structure of the human spliceosomal protein2 complex p14-sf3b155
55 c4nz3A_
not modelled
13.6
16
PDB header: hydrolaseChain: A: PDB Molecule: deacetylase da1;PDBTitle: structure of vibrio cholerae chitin de-n-acetylase in complex with2 di(n-acetyl-d-glucosamine) (cbs) in p 21 21 21
56 d2ceva_
not modelled
13.5
16
Fold: Arginase/deacetylaseSuperfamily: Arginase/deacetylaseFamily: Arginase-like amidino hydrolases
57 c3kwsB_
not modelled
13.5
12
PDB header: isomeraseChain: B: PDB Molecule: putative sugar isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
58 c3vylB_
not modelled
12.9
13
PDB header: isomeraseChain: B: PDB Molecule: l-ribulose 3-epimerase;PDBTitle: structure of l-ribulose 3-epimerase
59 c3cnyA_
not modelled
12.4
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: inositol catabolism protein iole;PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
60 c1uytC_
not modelled
12.1
10
PDB header: transferaseChain: C: PDB Molecule: acetyl-coa carboxylase;PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
61 d1mhhe_
not modelled
11.9
27
Fold: beta-Grasp (ubiquitin-like)Superfamily: Immunoglobulin-binding domainsFamily: Immunoglobulin-binding domains
62 c5zeeA_
not modelled
11.6
12
PDB header: hydrolase/inhibitorChain: A: PDB Molecule: arginase;PDBTitle: crystal structure of entamoeba histolytica arginase in complex with2 n(omega)-hydroxy-l-arginine (noha) at 1.74 a
63 c5l0lB_
not modelled
11.4
26
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein lpg0439
64 c4rtbA_
not modelled
10.8
15
PDB header: lyaseChain: A: PDB Molecule: hydg protein;PDBTitle: x-ray structure of the fefe-hydrogenase maturase hydg from2 carboxydothermus hydrogenoformans
65 d2aeba1
not modelled
10.5
19
Fold: Arginase/deacetylaseSuperfamily: Arginase/deacetylaseFamily: Arginase-like amidino hydrolases
66 d1jmla_
not modelled
10.5
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: Immunoglobulin-binding domainsFamily: Immunoglobulin-binding domains
67 c2f9jP_
not modelled
9.7
18
PDB header: rna binding proteinChain: P: PDB Molecule: splicing factor 3b subunit 1;PDBTitle: 3.0 angstrom resolution structure of a y22m mutant of the spliceosomal2 protein p14 bound to a region of sf3b155
68 d1yx1a1
not modelled
9.5
14
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: KguE-like
69 c4rl4B_
not modelled
9.3
23
PDB header: hydrolaseChain: B: PDB Molecule: gtp cyclohydrolase-2;PDBTitle: crystal structure of gtp cyclohydrolase ii from helicobacter pylori2 26695
70 c1gthD_
not modelled
9.3
22
PDB header: oxidoreductaseChain: D: PDB Molecule: dihydropyrimidine dehydrogenase;PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary complex with2 nadph and 5-iodouracil
71 c3nipB_
not modelled
9.3
13
PDB header: hydrolaseChain: B: PDB Molecule: 3-guanidinopropionase;PDBTitle: crystal structure of pseudomonas aeruginosa guanidinopropionase2 complexed with 1,6-diaminohexane
72 d1vioa1
not modelled
9.3
20
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase RsuA/RluD
73 c2qx2A_
not modelled
9.2
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sex pheromone staph-cam373;PDBTitle: structure of the c-terminal domain of sex pheromone staph-cam3732 precursor from staphylococcus aureus
74 d2glka1
not modelled
9.1
18
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
75 d1b25a2
not modelled
8.9
21
Fold: Aldehyde ferredoxin oxidoreductase, N-terminal domainSuperfamily: Aldehyde ferredoxin oxidoreductase, N-terminal domainFamily: Aldehyde ferredoxin oxidoreductase, N-terminal domain
76 d2gp4a1
not modelled
8.9
12
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: IlvD/EDD C-terminal domain-like
77 d2ptla_
not modelled
8.8
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: Immunoglobulin-binding domainsFamily: Immunoglobulin-binding domains
78 c1mk2B_
not modelled
8.8
29
PDB header: transcriptionChain: B: PDB Molecule: madh-interacting protein;PDBTitle: smad3 sbd complex
79 d1hz6a_
not modelled
8.7
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: Immunoglobulin-binding domainsFamily: Immunoglobulin-binding domains
80 d1k52a_
not modelled
8.6
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: Immunoglobulin-binding domainsFamily: Immunoglobulin-binding domains
81 c3pywA_
not modelled
8.5
8
PDB header: structural proteinChain: A: PDB Molecule: s-layer protein sap;PDBTitle: the structure of the slh domain from b. anthracis surface array2 protein at 1.8a
82 d1mo0a_
not modelled
8.4
20
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
83 c2htfA_
not modelled
8.3
50
PDB header: transferaseChain: A: PDB Molecule: dna polymerase mu;PDBTitle: the solution structure of the brct domain from human2 polymerase reveals homology with the tdt brct domain
84 c4nvtD_
not modelled
8.2
19
PDB header: isomeraseChain: D: PDB Molecule: triosephosphate isomerase;PDBTitle: crystal structure of triosephosphate isomerase from brucella2 melitensis
85 c3bdkB_
not modelled
8.0
23
PDB header: lyaseChain: B: PDB Molecule: d-mannonate dehydratase;PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
86 c4hn3C_
not modelled
7.9
18
PDB header: signaling proteinChain: C: PDB Molecule: lmo1757 protein;PDBTitle: the crystal structure of a sex pheromone precursor (lmo1757) from2 listeria monocytogenes egd-e
87 c3obeB_
not modelled
7.6
11
PDB header: isomeraseChain: B: PDB Molecule: sugar phosphate isomerase/epimerase;PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_3400)2 from parabacteroides distasonis atcc 8503 at 1.70 a resolution
88 d7reqb2
not modelled
7.6
29
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain
89 d2cyga1
not modelled
7.5
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
90 d1n55a_
not modelled
7.5
17
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
91 c4b8nC_
not modelled
7.3
6
PDB header: electron transportChain: C: PDB Molecule: cytochrome b5-host origin;PDBTitle: cytochrome b5 of ostreococcus tauri virus 2
92 d1oqya2
not modelled
7.2
33
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain
93 c2eivH_
not modelled
7.1
17
PDB header: hydrolaseChain: H: PDB Molecule: arginase;PDBTitle: crystal structure of the arginase from thermus thermophilus
94 d2a0ma1
not modelled
6.9
19
Fold: Arginase/deacetylaseSuperfamily: Arginase/deacetylaseFamily: Arginase-like amidino hydrolases
95 c2dp3A_
not modelled
6.9
17
PDB header: isomeraseChain: A: PDB Molecule: triosephosphate isomerase;PDBTitle: crystal structure of a double mutant (c202a/a198v) of triosephosphate2 isomerase from giardia lamblia
96 c3krsB_
not modelled
6.8
23
PDB header: isomeraseChain: B: PDB Molecule: triosephosphate isomerase;PDBTitle: structure of triosephosphate isomerase from cryptosporidium parvum at2 1.55a resolution
97 d1suxa_
not modelled
6.7
15
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
98 c4z3wB_
not modelled
6.7
14
PDB header: oxidoreductaseChain: B: PDB Molecule: benzoyl-coa reductase, putative;PDBTitle: active site complex bambc of benzoyl coenzyme a reductase in complex2 with 1,5 dienoyl-coa
99 d1xp3a1
not modelled
6.6
13
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Endonuclease IV