| 1 | c2vycA_
|
|
 |
100.0 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:biodegradative arginine decarboxylase;
PDBTitle: crystal structure of acid induced arginine decarboxylase2 from e. coli
|
|
|
|
| 2 | c5fkzE_
|
|
 |
100.0 |
25 |
PDB header:lyase
Chain: E: PDB Molecule:lysine decarboxylase, constitutive;
PDBTitle: structure of e.coli constitutive lysine decarboxylase
|
|
|
|
| 3 | c3n75E_
|
|
 |
100.0 |
25 |
PDB header:lyase
Chain: E: PDB Molecule:lysine decarboxylase, inducible;
PDBTitle: x-ray crystal structure of the escherichia coli inducible lysine2 decarboxylase ldci
|
|
|
|
| 4 | c1c4kA_
|
|
 |
100.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:protein (ornithine decarboxylase);
PDBTitle: ornithine decarboxylase mutant (gly121tyr)
|
|
|
|
| 5 | d1c4ka2
|
|
 |
100.0 |
25 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Ornithine decarboxylase major domain |
|
|
|
| 6 | c2x3lA_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:orn/lys/arg decarboxylase family protein;
PDBTitle: crystal structure of the orn_lys_arg decarboxylase family2 protein sar0482 from methicillin-resistant staphylococcus3 aureus
|
|
|
|
| 7 | d1rv3a_
|
|
 |
100.0 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
|
| 8 | d1bj4a_
|
|
 |
100.0 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
|
| 9 | c6cd1A_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of medicago truncatula serine2 hydroxymethyltransferase 3 (mtshmt3), complexes with reaction3 intermediates
|
|
|
|
| 10 | d1ejia_
|
|
 |
100.0 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
|
| 11 | d2z67a1
|
|
 |
100.0 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:SepSecS-like |
|
|
|
| 12 | d2a7va1
|
|
 |
100.0 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
|
| 13 | c2a7vA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: human mitochondrial serine hydroxymethyltransferase 2
|
|
|
|
| 14 | c4o6zC_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of serine hydroxymethyltransferase with covalently2 bound plp schiff-base from plasmodium falciparum
|
|
|
|
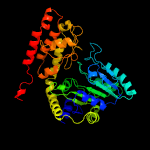
| 15 | c3h7fB_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:serine hydroxymethyltransferase 1;
PDBTitle: crystal structure of serine hydroxymethyltransferase from2 mycobacterium tuberculosis
|
|
|
|
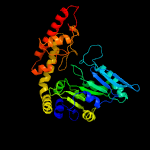
| 16 | c5z0yA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystallization and structure determination of cytoplasm serine2 hydroxymethyltransferase (shmt) from pichia pastoris
|
|
|
|
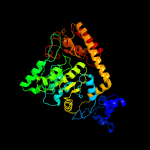
| 17 | c3hl2D_
|
|
 |
100.0 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:o-phosphoseryl-trna(sec) selenium transferase;
PDBTitle: the crystal structure of the human sepsecs-trnasec complex
|
|
|
|
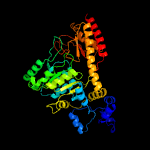
| 18 | d3bc8a1
|
|
 |
100.0 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:SepSecS-like |
|
|
|
| 19 | c3ecdC_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:serine hydroxymethyltransferase 2;
PDBTitle: crystal structure of serine hydroxymethyltransferase from burkholderia2 pseudomallei
|
|
|
|
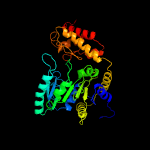
| 20 | c4wxfC_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of l-serine hydroxymethyltransferase in complex with2 glycine
|
|
|
|
| 21 | c4q6rB_ |
|
not modelled |
100.0 |
14 |
PDB header:lyase/lyase inhibitor
Chain: B: PDB Molecule:sphingosine-1-phosphate lyase 1;
PDBTitle: crystal structure of human sphingosine-1-phosphate lyase in complex2 with inhibitor 6-[(2r)-4-(4-benzyl-7-chlorophthalazin-1-yl)-2-3 methylpiperazin-1-yl]pyridine-3-carbonitrile
|
|
|
| 22 | c3n0lA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of serine hydroxymethyltransferase from2 campylobacter jejuni
|
|
|
| 23 | c5vc2A_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of a serine hydroxymethyltransferase from2 helicobacter pylori
|
|
|
| 24 | d1dfoa_ |
|
not modelled |
100.0 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 25 | c4j5uB_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: x-ray crystal structure of a serine hydroxymethyltransferase with2 covalently bound plp from rickettsia rickettsii str. sheila smith
|
|
|
| 26 | c4bheI_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: I: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: methanococcus jannaschii serine hydroxymethyl-transferase2 in complex with plp
|
|
|
| 27 | d1kl1a_ |
|
not modelled |
99.9 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 28 | c2dkjB_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of t.th.hb8 serine hydroxymethyltransferase
|
|
|
| 29 | d2e7ja1 |
|
not modelled |
99.9 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:SepSecS-like |
|
|
| 30 | c4n0wA_ |
|
not modelled |
99.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: x-ray crystal structure of a serine hydroxymethyltransferase from2 burkholderia cenocepacia with covalently attached pyridoxal phosphate
|
|
|
| 31 | c6hrhA_ |
|
not modelled |
99.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:5-aminolevulinate synthase, erythroid-specific,
PDBTitle: structure of human erythroid-specific 5'-aminolevulinate synthase,2 alas2
|
|
|
| 32 | c5k1rB_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:burkholderia pseudomallei sphingosine-1-phosphate lyase
PDBTitle: structure of burkholderia pseudomallei k96243 sphingosine-1-phosphate2 lyase bpss2021
|
|
|
| 33 | c3madA_ |
|
not modelled |
99.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:sphingosine-1-phosphate lyase;
PDBTitle: crystal structure of stspl (symmetric form)
|
|
|
| 34 | c3mafB_ |
|
not modelled |
99.9 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:sphingosine-1-phosphate lyase;
PDBTitle: crystal structure of stspl (asymmetric form)
|
|
|
| 35 | c3mc6C_ |
|
not modelled |
99.9 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:sphingosine-1-phosphate lyase;
PDBTitle: crystal structure of scdpl1
|
|
|
| 36 | d1vjoa_ |
|
not modelled |
99.9 |
20 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
|
|
| 37 | c5txtA_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:5-aminolevulinate synthase, mitochondrial;
PDBTitle: structure of asymmetric apo/holo alas dimer from s. cerevisiae
|
|
|
| 38 | d1wyua1 |
|
not modelled |
99.9 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Glycine dehydrogenase subunits (GDC-P) |
|
|
| 39 | d1wyub1 |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Glycine dehydrogenase subunits (GDC-P) |
|
|
| 40 | c3f9tB_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:l-tyrosine decarboxylase mfna;
PDBTitle: crystal structure of l-tyrosine decarboxylase mfna (ec 4.1.1.25)2 (np_247014.1) from methanococcus jannaschii at 2.11 a resolution
|
|
|
| 41 | c5o5cD_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:putative decarboxylase involved in desferrioxamine
PDBTitle: the crystal structure of dfoj, the desferrioxamine biosynthetic2 pathway lysine decarboxylase from the fire blight disease pathogen3 erwinia amylovora
|
|
|
| 42 | c6ewqA_ |
|
not modelled |
99.9 |
15 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative capsular polysaccharide biosynthesis protein;
PDBTitle: putative sugar aminotransferase spr1654 from streptococcus pneumoniae,2 plp-form
|
|
|
| 43 | d1js3a_ |
|
not modelled |
99.9 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Pyridoxal-dependent decarboxylase |
|
|
| 44 | c4ritB_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:pyridoxal-dependent decarboxylase;
PDBTitle: the yellow crystal structure of pyridoxal-dependent decarboxylase from2 sphaerobacter thermophilus dsm 20745
|
|
|
| 45 | c4obuG_ |
|
not modelled |
99.9 |
16 |
PDB header:lyase
Chain: G: PDB Molecule:pyridoxal-dependent decarboxylase domain protein;
PDBTitle: ruminococcus gnavus tryptophan decarboxylase rumgna_01526 (apo)
|
|
|
| 46 | d1m6sa_ |
|
not modelled |
99.9 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
|
|
| 47 | c2jisA_ |
|
not modelled |
99.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:cysteine sulfinic acid decarboxylase;
PDBTitle: human cysteine sulfinic acid decarboxylase (csad) in2 complex with plp.
|
|
|
| 48 | c3nnkC_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:ureidoglycine-glyoxylate aminotransferase;
PDBTitle: biochemical and structural characterization of a ureidoglycine2 aminotransferase in the klebsiella pneumoniae uric acid catabolic3 pathway
|
|
|
| 49 | c2z9wA_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:aspartate aminotransferase;
PDBTitle: crystal structure of pyridoxamine-pyruvate aminotransferase complexed2 with pyridoxal
|
|
|
| 50 | c2w8wA_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:serine palmitoyltransferase;
PDBTitle: n100y spt with plp-ser
|
|
|
| 51 | c3bcxA_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:cdp-6-deoxy-l-threo-d-glycero-4-hexulose-3-dehydrase;
PDBTitle: e1 dehydrase
|
|
|
| 52 | d1h0ca_ |
|
not modelled |
99.9 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
|
|
| 53 | d1o69a_ |
|
not modelled |
99.8 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 54 | c2hzpA_ |
|
not modelled |
99.8 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynureninase;
PDBTitle: crystal structure of homo sapiens kynureninase
|
|
|
| 55 | c5utsC_ |
|
not modelled |
99.8 |
10 |
PDB header:lyase
Chain: C: PDB Molecule:c-s lyase egt2;
PDBTitle: carbon sulfoxide lyase, egt2 in the ergothioneine biosynthesis pathway
|
|
|
| 56 | c4e1oC_ |
|
not modelled |
99.8 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:histidine decarboxylase;
PDBTitle: human histidine decarboxylase complex with histidine methyl ester2 (hme)
|
|
|
| 57 | c6jrlA_ |
|
not modelled |
99.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3,4-dihydroxyphenylacetaldehyde synthase;
PDBTitle: crystal structure of drosophila alpha methyldopa-resistant protein/3,2 4-dihydroxyphenylacetaldehyde synthase
|
|
|
| 58 | c3e9kA_ |
|
not modelled |
99.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynureninase;
PDBTitle: crystal structure of homo sapiens kynureninase-3-hydroxyhippuric acid2 inhibitor complex
|
|
|
| 59 | c6eewC_ |
|
not modelled |
99.8 |
19 |
PDB header:lyase
Chain: C: PDB Molecule:aromatic-l-amino-acid decarboxylase;
PDBTitle: crystal structure of catharanthus roseus tryptophan decarboxylase in2 complex with l-tryptophan
|
|
|
| 60 | c3l8aB_ |
|
not modelled |
99.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:putative aminotransferase, probable beta-cystathionase;
PDBTitle: crystal structure of metc from streptococcus mutans
|
|
|
| 61 | c3hbxB_ |
|
not modelled |
99.8 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:glutamate decarboxylase 1;
PDBTitle: crystal structure of gad1 from arabidopsis thaliana
|
|
|
| 62 | c2po3B_ |
|
not modelled |
99.8 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:4-dehydrase;
PDBTitle: crystal structure analysis of desi in the presence of its2 tdp-sugar product
|
|
|
| 63 | c4qgrA_ |
|
not modelled |
99.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:degt/dnrj/eryc1/strs aminotransferase;
PDBTitle: crystal structure of a degt dnrj eryc1 strs aminotransferase from2 brucella abortus
|
|
|
| 64 | c6eeqA_ |
|
not modelled |
99.8 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxyphenylacetaldehyde synthase;
PDBTitle: crystal structure of rhodiola rosea 4-hydroxyphenylacetaldehyde2 synthase
|
|
|
| 65 | c3islA_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:purine catabolism protein pucg;
PDBTitle: crystal structure of ureidoglycine-glyoxylate aminotransferase (pucg)2 from bacillus subtilis
|
|
|
| 66 | d1tpla_ |
|
not modelled |
99.8 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
|
|
| 67 | c5k8bA_ |
|
not modelled |
99.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:8-amino-3,8-dideoxy-alpha-d-manno-octulosonate
PDBTitle: x-ray structure of kdna, 8-amino-3,8-dideoxy-alpha-d-manno-2 octulosonate transaminase, from shewanella oneidensis in the presence3 of the external aldimine with plp and glutamate
|
|
|
| 68 | c6enzA_ |
|
not modelled |
99.8 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:acidic amino acid decarboxylase gadl1;
PDBTitle: crystal structure of mouse gadl1
|
|
|
| 69 | d1mdoa_ |
|
not modelled |
99.8 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 70 | c5gp4C_ |
|
not modelled |
99.8 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:glutamate decarboxylase;
PDBTitle: lactobacillus brevis cgmcc 1306 glutamate decarboxylase
|
|
|
| 71 | c3a2bA_ |
|
not modelled |
99.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:serine palmitoyltransferase;
PDBTitle: crystal structure of serine palmitoyltransferase from sphingobacterium2 multivorum with substrate l-serine
|
|
|
| 72 | c3wgcB_ |
|
not modelled |
99.8 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:l-allo-threonine aldolase;
PDBTitle: aeromonas jandaei l-allo-threonine aldolase h128y/s292r double mutant
|
|
|
| 73 | d2v1pa1 |
|
not modelled |
99.8 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
|
|
| 74 | d1v72a1 |
|
not modelled |
99.8 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
|
|
| 75 | c3hqtB_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:cai-1 autoinducer synthase;
PDBTitle: plp-dependent acyl-coa transferase cqsa
|
|
|
| 76 | c2r0tA_ |
|
not modelled |
99.8 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:pyridoxamine 5-phosphate-dependent dehydrase;
PDBTitle: crystal structure of gdp-4-keto-6-deoxymannose-3-dehydratase with a2 trapped plp-glutamate geminal diamine
|
|
|
| 77 | d1fc4a_ |
|
not modelled |
99.8 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 78 | c4lnjA_ |
|
not modelled |
99.8 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:low-specificity l-threonine aldolase;
PDBTitle: structure of escherichia coli threonine aldolase in unliganded form
|
|
|
| 79 | c3k40B_ |
|
not modelled |
99.8 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:aromatic-l-amino-acid decarboxylase;
PDBTitle: crystal structure of drosophila 3,4-dihydroxyphenylalanine2 decarboxylase
|
|
|
| 80 | d2ch1a1 |
|
not modelled |
99.8 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
|
|
| 81 | c2ogeC_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:transaminase;
PDBTitle: x-ray structure of s. venezuelae desv in its internal2 aldimine form
|
|
|
| 82 | c3zrrB_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:serine-pyruvate aminotransferase (agxt);
PDBTitle: crystal structure and substrate specificity of a thermophilic2 archaeal serine : pyruvate aminotransferase from sulfolobus3 solfataricus
|
|
|
| 83 | c2dr1A_ |
|
not modelled |
99.8 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:386aa long hypothetical serine aminotransferase;
PDBTitle: crystal structure of the ph1308 protein from pyrococcus horikoshii ot3
|
|
|
| 84 | c3tqxA_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-3-ketobutyrate coenzyme a ligase;
PDBTitle: structure of the 2-amino-3-ketobutyrate coenzyme a ligase (kbl) from2 coxiella burnetii
|
|
|
| 85 | d2bwna1 |
|
not modelled |
99.8 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 86 | c3wy7D_ |
|
not modelled |
99.8 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:8-amino-7-oxononanoate synthase;
PDBTitle: crystal structure of mycobacterium smegmatis 7-keto-8-aminopelargonic2 acid (kapa) synthase biof
|
|
|
| 87 | c3uwcA_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:nucleotide-sugar aminotransferase;
PDBTitle: structure of an aminotransferase (degt-dnrj-eryc1-strs family) from2 coxiella burnetii in complex with pmp
|
|
|
| 88 | d1bs0a_ |
|
not modelled |
99.8 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 89 | c3nysA_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase wbpe;
PDBTitle: x-ray structure of the k185a mutant of wbpe (wlbe) from pseudomonas2 aeruginosa in complex with plp at 1.45 angstrom resolution
|
|
|
| 90 | c2c7tA_ |
|
not modelled |
99.8 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:glutamine-2-deoxy-scyllo-inosose aminotransferase;
PDBTitle: crystal structure of the plp-bound form of btrr, a dual functional2 aminotransferase involved in butirosin biosynthesis.
|
|
|
| 91 | c3w1hB_ |
|
not modelled |
99.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:l-seryl-trna(sec) selenium transferase;
PDBTitle: crystal structure of the selenocysteine synthase sela from aquifex2 aeolicus
|
|
|
| 92 | c3ffrA_ |
|
not modelled |
99.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoserine aminotransferase serc;
PDBTitle: crystal structure of a phosphoserine aminotransferase serc (chu_0995)2 from cytophaga hutchinsonii atcc 33406 at 1.75 a resolution
|
|
|
| 93 | c5w70B_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:l-glutamine:2-deoxy-scyllo-inosose aminotransferase;
PDBTitle: x-ray structure of rbmb from streptomyces ribosidificus
|
|
|
| 94 | d1c7na_ |
|
not modelled |
99.8 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
|
|
| 95 | c2huuA_ |
|
not modelled |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:alanine glyoxylate aminotransferase;
PDBTitle: crystal structure of aedes aegypti alanine glyoxylate aminotransferase2 in complex with alanine
|
|
|
| 96 | c6eeiA_ |
|
not modelled |
99.8 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:tyrosine decarboxylase 1;
PDBTitle: crystal structure of arabidopsis thaliana phenylacetaldehyde synthase2 in complex with l-phenylalanine
|
|
|
| 97 | d1svva_ |
|
not modelled |
99.8 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
|
|
| 98 | d2aeua1 |
|
not modelled |
99.8 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:SelA-like |
|
|
| 99 | c3kaxB_ |
|
not modelled |
99.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:aminotransferase, classes i and ii;
PDBTitle: crystal structure of a putative c-s lyase from bacillus anthracis
|
|
|
| 100 | c3b1dD_ |
|
not modelled |
99.8 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:betac-s lyase;
PDBTitle: crystal structure of betac-s lyase from streptococcus anginosus in2 complex with l-serine: external aldimine form
|
|
|
| 101 | c3frkB_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:qdtb;
PDBTitle: x-ray structure of qdtb from t. thermosaccharolyticum in2 complex with a plp:tdp-3-aminoquinovose aldimine
|
|
|
| 102 | c3ke3A_ |
|
not modelled |
99.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative serine-pyruvate aminotransferase;
PDBTitle: crystal structure of putative serine-pyruvate aminotransferase2 (yp_263484.1) from psychrobacter arcticum 273-4 at 2.20 a resolution
|
|
|
| 103 | d1c4ka3 |
|
not modelled |
99.8 |
19 |
Fold:Ornithine decarboxylase C-terminal domain
Superfamily:Ornithine decarboxylase C-terminal domain
Family:Ornithine decarboxylase C-terminal domain |
|
|
| 104 | c5jayB_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:8-amino-7-oxononanoate synthase;
PDBTitle: crystal structure of an 8-amino-7-oxononanoate synthase from2 burkholderia xenovorans
|
|
|
| 105 | c3pj0D_ |
|
not modelled |
99.8 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:lmo0305 protein;
PDBTitle: crystal structure of a putative l-allo-threonine aldolase (lmo0305)2 from listeria monocytogenes egd-e at 1.80 a resolution
|
|
|
| 106 | c3f0hA_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase;
PDBTitle: crystal structure of aminotransferase (rer070207000802) from2 eubacterium rectale at 1.70 a resolution
|
|
|
| 107 | c3dr4B_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:putative perosamine synthetase;
PDBTitle: gdp-perosamine synthase k186a mutant from caulobacter2 crescentus with bound sugar ligand
|
|
|
| 108 | c4q76B_ |
|
not modelled |
99.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:cysteine desulfurase 2, chloroplastic;
PDBTitle: crystal structure of nfs2 c384s mutant, the plastidial cysteine2 desulfurase from arabidopsis thaliana
|
|
|
| 109 | c6eemB_ |
|
not modelled |
99.8 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:tyrosine/dopa decarboxylase;
PDBTitle: crystal structure of papaver somniferum tyrosine decarboxylase in2 complex with l-tyrosine
|
|
|
| 110 | c2okkA_ |
|
not modelled |
99.8 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:glutamate decarboxylase 2;
PDBTitle: the x-ray crystal structure of the 65kda isoform of glutamic acid2 decarboxylase (gad65)
|
|
|
| 111 | c3ju7B_ |
|
not modelled |
99.8 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:putative plp-dependent aminotransferase;
PDBTitle: crystal structure of putative plp-dependent aminotransferase2 (np_978343.1) from bacillus cereus atcc 10987 at 2.19 a resolution
|
|
|
| 112 | c2cb1A_ |
|
not modelled |
99.8 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:o-acetyl homoserine sulfhydrylase;
PDBTitle: crystal structure of o-actetyl homoserine sulfhydrylase2 from thermus thermophilus hb8,oah2.
|
|
|
| 113 | d1qgna_ |
|
not modelled |
99.7 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
|
|
| 114 | d1c7ga_ |
|
not modelled |
99.7 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
|
|
| 115 | d1jf9a_ |
|
not modelled |
99.7 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
|
|
| 116 | c3op7A_ |
|
not modelled |
99.7 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase class i and ii;
PDBTitle: crystal structure of a plp-dependent aminotransferase (zp_03625122.1)2 from streptococcus suis 89-1591 at 1.70 a resolution
|
|
|
| 117 | c1d2fB_ |
|
not modelled |
99.7 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:maly protein;
PDBTitle: x-ray structure of maly from escherichia coli: a pyridoxal-5'-2 phosphate-dependent enzyme acting as a modulator in mal gene3 expression
|
|
|
| 118 | d1b9ha_ |
|
not modelled |
99.7 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 119 | c4ytjC_ |
|
not modelled |
99.7 |
12 |
PDB header:transferase
Chain: C: PDB Molecule:cals13;
PDBTitle: crystal structure of sugar aminotransferase cals13 from micromonospora2 echinospora
|
|
|
| 120 | c5vyeA_ |
|
not modelled |
99.7 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:l-threonine aldolase;
PDBTitle: crystal structure of l-threonine aldolase from pseudomonas putida
|
|
|