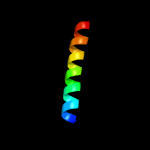1 c1coiA_
78.6
52
PDB header: alpha-helical bundleChain: A: PDB Molecule: coil-vald;PDBTitle: designed trimeric coiled coil-vald
2 c3cvfA_
61.0
25
PDB header: signaling proteinChain: A: PDB Molecule: homer protein homolog 3;PDBTitle: crystal structure of the carboxy terminus of homer3
3 c6h9mA_
50.6
26
PDB header: membrane proteinChain: A: PDB Molecule: coiled-coil domain-containing protein 90b, mitochondrial,PDBTitle: coiled-coil domain-containing protein 90b residues 43-125 from homo2 sapiens fused to a gcn4 adaptor
4 c5xauC_
43.2
17
PDB header: cell adhesionChain: C: PDB Molecule: laminin subunit gamma-1;PDBTitle: crystal structure of integrin binding fragment of laminin-511
5 c6dmpA_
37.3
44
PDB header: de novo proteinChain: A: PDB Molecule: designed orthogonal protein dhd13_xaaa_a;PDBTitle: de novo design of a protein heterodimer with specificity mediated by2 hydrogen bond networks
6 c5uxtB_
35.9
48
PDB header: de novo proteinChain: B: PDB Molecule: coiled-coil trimer with glu:trp:lys triad;PDBTitle: coiled-coil trimer with glu:trp:lys triad
7 c4widA_
34.6
23
PDB header: viral proteinChain: A: PDB Molecule: rhul123;PDBTitle: crystal structure of the immediate-early 1 protein (ie1) at 2.312 angstrom (tetragonal form after crystal dehydration)
8 c5uxtC_
34.6
48
PDB header: de novo proteinChain: C: PDB Molecule: coiled-coil trimer with glu:trp:lys triad;PDBTitle: coiled-coil trimer with glu:trp:lys triad
9 c5uxtA_
33.9
48
PDB header: de novo proteinChain: A: PDB Molecule: coiled-coil trimer with glu:trp:lys triad;PDBTitle: coiled-coil trimer with glu:trp:lys triad
10 c3sjrB_
29.0
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of conserved unkown function protein cv_1783 from2 chromobacterium violaceum atcc 12472
11 c2pnvA_
25.7
30
PDB header: membrane proteinChain: A: PDB Molecule: small conductance calcium-activated potassiumPDBTitle: crystal structure of the leucine zipper domain of small-2 conductance ca2+-activated k+ (skca) channel from rattus3 norvegicus
12 c4zmhA_
23.2
40
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a five-domain gh115 alpha-glucuronidase from the2 marine bacterium saccharophagus degradans 2-40t
13 c1cosC_
20.7
40
PDB header: alpha-helical bundleChain: C: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-helical bundle
14 c1cosB_
20.7
40
PDB header: alpha-helical bundleChain: B: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-helical bundle
15 c1cosA_
20.7
40
PDB header: alpha-helical bundleChain: A: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-helical bundle
16 c3swyB_
20.1
58
PDB header: transport proteinChain: B: PDB Molecule: cyclic nucleotide-gated cation channel alpha-3;PDBTitle: cnga3 626-672 containing clz domain
17 c2a93B_
19.6
41
PDB header: leucine zippersChain: B: PDB Molecule: c-myc-max heterodimeric leucine zipper;PDBTitle: nmr solution structure of the c-myc-max heterodimeric2 leucine zipper, 40 structures
18 c5k92C_
17.3
40
PDB header: de novo proteinChain: C: PDB Molecule: apo-(csl16c)3;PDBTitle: crystal structure of an apo tris-thiolate binding site in a de novo2 three stranded coiled coil peptide
19 c5k92B_
17.3
40
PDB header: de novo proteinChain: B: PDB Molecule: apo-(csl16c)3;PDBTitle: crystal structure of an apo tris-thiolate binding site in a de novo2 three stranded coiled coil peptide
20 c5k92A_
17.3
40
PDB header: de novo proteinChain: A: PDB Molecule: apo-(csl16c)3;PDBTitle: crystal structure of an apo tris-thiolate binding site in a de novo2 three stranded coiled coil peptide
21 c5by3A_
not modelled
17.2
27
PDB header: sugar binding proteinChain: A: PDB Molecule: btgh115a;PDBTitle: a novel family gh115 4-o-methyl-alpha-glucuronidase, btgh115a, with2 specificity for decorated arabinogalactans
22 c4u5tB_
not modelled
16.8
35
PDB header: transcription/transcription inhibitorChain: B: PDB Molecule: vbp leucine zipper;PDBTitle: crystal structure of vbp leucine zipper with bound arylstibonic acid
23 c4e18B_
not modelled
16.3
30
PDB header: cell adhesionChain: B: PDB Molecule: catenin alpha-1;PDBTitle: alpha-e-catenin is an autoinhibited molecule that co-activates2 vinculin
24 c2jgoA_
not modelled
16.2
40
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l9c;PDBTitle: structure of the arsenated de novo designed peptide coil ser l9c
25 c2jgoC_
not modelled
16.2
40
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l9c;PDBTitle: structure of the arsenated de novo designed peptide coil ser l9c
26 c2jgoB_
not modelled
16.2
40
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l9c;PDBTitle: structure of the arsenated de novo designed peptide coil ser l9c
27 c3ljmA_
not modelled
16.2
40
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l9c;PDBTitle: structure of de novo designed apo peptide coil ser l9c
28 c3ljmC_
not modelled
16.2
40
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l9c;PDBTitle: structure of de novo designed apo peptide coil ser l9c
29 c3ljmB_
not modelled
16.2
40
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l9c;PDBTitle: structure of de novo designed apo peptide coil ser l9c
30 c3iynR_
not modelled
15.4
18
PDB header: virusChain: R: PDB Molecule: hexon-associated protein;PDBTitle: 3.6-angstrom cryoem structure of human adenovirus type 5
31 c2wpyA_
not modelled
14.8
35
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with one vxxnxxx motif2 coordinating chloride
32 c4g1aC_
not modelled
14.3
38
PDB header: metal binding proteinChain: C: PDB Molecule: aq-c16c19 peptide;PDBTitle: metal-binding properties of a self-assembled coiled coil: formation of2 a polynuclear cd-thiolated cluster
33 c6a9wA_
not modelled
14.2
27
PDB header: replicationChain: A: PDB Molecule: primase;PDBTitle: structure of the bifunctional dna primase-polymerase from phage nrs-1
34 c2oqqB_
not modelled
14.2
43
PDB header: transcriptionChain: B: PDB Molecule: transcription factor hy5;PDBTitle: crystal structure of hy5 leucine zipper homodimer from arabidopsis2 thaliana
35 c3w8vC_
not modelled
14.1
35
PDB header: transcriptionChain: C: PDB Molecule: gcn4n coiled coil peptide;PDBTitle: crystal structure analysis of the synthetic gcn4 coiled coil peptide
36 c3w8vB_
not modelled
14.1
35
PDB header: transcriptionChain: B: PDB Molecule: gcn4n coiled coil peptide;PDBTitle: crystal structure analysis of the synthetic gcn4 coiled coil peptide
37 c3w8vA_
not modelled
14.1
35
PDB header: transcriptionChain: A: PDB Molecule: gcn4n coiled coil peptide;PDBTitle: crystal structure analysis of the synthetic gcn4 coiled coil peptide
38 c5kb0A_
not modelled
14.1
37
PDB header: de novo proteinChain: A: PDB Molecule: pb(ii)zn(ii)(grand coil ser-l16cl30h)3+;PDBTitle: crystal structure of a tris-thiolate pb(ii) complex in a de novo2 three-stranded coiled coil peptide
39 c1ztaA_
not modelled
13.5
34
PDB header: dna-binding motifChain: A: PDB Molecule: leucine zipper monomer;PDBTitle: the solution structure of a leucine-zipper motif peptide
40 c1ce0B_
not modelled
12.3
25
PDB header: hiv-1 envelope proteinChain: B: PDB Molecule: protein (leucine zipper model h38-p1);PDBTitle: trimerization specificity in hiv-1 gp41: analysis with a2 gcn4 leucine zipper model
41 c1ij2C_
not modelled
12.3
32
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
42 c2hw2A_
not modelled
12.0
28
PDB header: transferaseChain: A: PDB Molecule: rifampin adp-ribosyl transferase;PDBTitle: crystal structure of rifampin adp-ribosyl transferase in complex with2 rifampin
43 c1rb6C_
not modelled
11.9
32
PDB header: dna binding proteinChain: C: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as n16a2 tetragonal form
44 c3k7zA_
not modelled
11.9
32
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
45 c1rb1B_
not modelled
11.9
32
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
46 c1rb1A_
not modelled
11.9
32
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
47 c1swiA_
not modelled
11.9
32
PDB header: leucine zipperChain: A: PDB Molecule: gcn4p1;PDBTitle: gcn4-leucine zipper core mutant as n16a complexed with benzene
48 c3k7zB_
not modelled
11.9
32
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
49 d1vkoa1
not modelled
11.7
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
50 c1ij3C_
not modelled
11.7
32
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
51 c1ij3B_
not modelled
11.7
32
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
52 c1ij2B_
not modelled
11.5
32
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
53 c3vgxC_
not modelled
11.4
27
PDB header: membrane proteinChain: C: PDB Molecule: envelope glycoprotein gp160;PDBTitle: structure of gp41 t21/cp621-652
54 c4cgbE_
not modelled
11.0
33
PDB header: cell cycleChain: E: PDB Molecule: echinoderm microtubule-associated protein-like 2;PDBTitle: crystal structure of the trimerization domain of eml2
55 c5cffD_
not modelled
11.0
29
PDB header: transcription/rna binding proteinChain: D: PDB Molecule: miranda;PDBTitle: crystal structure of miranda/staufen dsrbd5 complex
56 c5ep6C_
not modelled
11.0
20
PDB header: protein binding/transferaseChain: C: PDB Molecule: 5-azacytidine-induced protein 2;PDBTitle: the crystal structure of nap1 in complex with tbk1
57 c1lq7A_
not modelled
10.8
50
PDB header: de novo proteinChain: A: PDB Molecule: alpha3w;PDBTitle: de novo designed protein model of radical enzymes
58 c5ep6A_
not modelled
10.7
20
PDB header: protein binding/transferaseChain: A: PDB Molecule: 5-azacytidine-induced protein 2;PDBTitle: the crystal structure of nap1 in complex with tbk1
59 c2mi7A_
not modelled
10.7
50
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein a3y;PDBTitle: solution nmr structure of alpha3y
60 c2lxyA_
not modelled
10.7
50
PDB header: de novo proteinChain: A: PDB Molecule: 2-mercaptophenol-alpha3c;PDBTitle: nmr structure of 2-mercaptophenol-alpha3c
61 c2o7hF_
not modelled
10.4
32
PDB header: transcriptionChain: F: PDB Molecule: general control protein gcn4;PDBTitle: crystal structure of trimeric coiled coil gcn4 leucine zipper
62 c2ql2A_
not modelled
9.8
60
PDB header: transcription/dnaChain: A: PDB Molecule: transcription factor e2-alpha;PDBTitle: crystal structure of the basic-helix-loop-helix domains of2 the heterodimer e47/neurod1 bound to dna
63 c4cgbA_
not modelled
9.6
33
PDB header: cell cycleChain: A: PDB Molecule: echinoderm microtubule-associated protein-like 2;PDBTitle: crystal structure of the trimerization domain of eml2
64 d2p4ka2
not modelled
9.3
33
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain
65 c4bl6A_
not modelled
9.2
14
PDB header: protein transportChain: A: PDB Molecule: protein bicaudal d;PDBTitle: bicaudal-d uses a parallel, homodimeric coiled coil with heterotypic2 registry to co-ordinate recruitment of cargos to dynein
66 c6gapB_
not modelled
8.8
16
PDB header: viral proteinChain: B: PDB Molecule: outer capsid protein sigma-1;PDBTitle: crystal structure of the t3d reovirus sigma1 coiled coil tail and body
67 c2x6pC_
not modelled
8.8
36
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l19c;PDBTitle: crystal structure of coil ser l19c
68 c2x6pA_
not modelled
8.8
36
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l19c;PDBTitle: crystal structure of coil ser l19c
69 c2x6pB_
not modelled
8.8
36
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l19c;PDBTitle: crystal structure of coil ser l19c
70 c6gbrA_
not modelled
8.4
31
PDB header: viral proteinChain: A: PDB Molecule: polymerase cofactor vp35;PDBTitle: crystal structure of the oligomerization domain of vp35 from reston2 virus, mercury derivative
71 c1htmB_
not modelled
8.3
17
PDB header: viral proteinChain: B: PDB Molecule: hemagglutinin ha2 chain;PDBTitle: structure of influenza haemagglutinin at the ph of membrane2 fusion
72 c3cveC_
not modelled
8.1
21
PDB header: signaling proteinChain: C: PDB Molecule: homer protein homolog 1;PDBTitle: crystal structure of the carboxy terminus of homer1
73 c6fueC_
not modelled
7.7
64
PDB header: protein transportChain: C: PDB Molecule: fapf;PDBTitle: periplasmic coiled coil domain of the fapf amyloid transporter
74 c3mtuD_
not modelled
7.3
18
PDB header: contractile proteinChain: D: PDB Molecule: tropomyosin alpha-1 chain,microtubule-associated proteinPDBTitle: structure of the tropomyosin overlap complex from chicken smooth2 muscle
75 c4iffC_
not modelled
7.1
21
PDB header: cell cycleChain: C: PDB Molecule: fusion of phage phi29 gp7 protein and cell division proteinPDBTitle: structural organization of ftsb, a transmembrane protein of the2 bacterial divisome
76 c1m7lA_
not modelled
7.0
40
PDB header: sugar binding proteinChain: A: PDB Molecule: pulmonary surfactant-associated protein d;PDBTitle: solution structure of the coiled-coil trimerization domain2 from lung surfactant protein d
77 c5eofB_
not modelled
6.8
15
PDB header: protein binding/transferaseChain: B: PDB Molecule: optineurin;PDBTitle: crystal structure of optn ntd and tbk1 ctd complex
78 c1kwwC_
not modelled
6.6
13
PDB header: immune system, sugar binding proteinChain: C: PDB Molecule: mannose-binding protein a;PDBTitle: rat mannose protein a complexed with a-me-fuc.
79 d1p1ja1
not modelled
6.5
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
80 c3h3mB_
not modelled
6.4
7
PDB header: structural genomicsChain: B: PDB Molecule: flagellar protein flit;PDBTitle: crystal structure of flagellar protein flit from bordetella2 bronchiseptica
81 c2rukA_
not modelled
6.4
50
PDB header: transcriptionChain: A: PDB Molecule: cellular tumor antigen p53;PDBTitle: solution structure of the complex between p53 transactivation domain 22 and tfiih p62 ph domain
82 c1qceB_
not modelled
6.3
47
PDB header: viral proteinChain: B: PDB Molecule: protein (gp41);PDBTitle: solution nmr structure of ectodomain of siv gp41,2 restrained regularized mean structure plus 29 simulated3 annealing structures
83 d1b06a2
not modelled
6.3
25
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain
84 c6gboG_
not modelled
6.3
34
PDB header: viral proteinChain: G: PDB Molecule: polymerase cofactor vp35;PDBTitle: crystal structure of the oligomerization domain of vp35 from ebola2 virus
85 c6e6aB_
not modelled
6.0
18
PDB header: protein bindingChain: B: PDB Molecule: inclusion membrane protein a;PDBTitle: triclinic crystal form of inca g144a point mutant
86 c1iojA_
not modelled
5.9
36
PDB header: apolipoproteinChain: A: PDB Molecule: apoc-i;PDBTitle: human apolipoprotein c-i, nmr, 18 structures
87 c3w93C_
not modelled
5.9
58
PDB header: transcriptionChain: C: PDB Molecule: coiled coil peptide;PDBTitle: crystal structure analysis of the synthetic gcn4 ester coiled coil2 peptide
88 c2yfaA_
not modelled
5.8
21
PDB header: receptorChain: A: PDB Molecule: methyl-accepting chemotaxis transducer;PDBTitle: x-ray structure of mcps ligand binding domain in complex with malate
89 c3h5fB_
not modelled
5.8
58
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l16l-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
90 c5u9uA_
not modelled
5.8
58
PDB header: de novo proteinChain: A: PDB Molecule: apo-(coilser l16(dcy))3;PDBTitle: de novo three-stranded coiled coil peptide containing a tris-thiolate2 site engineered by d-cysteine ligands
91 c5u9uC_
not modelled
5.8
58
PDB header: de novo proteinChain: C: PDB Molecule: apo-(coilser l16(dcy))3;PDBTitle: de novo three-stranded coiled coil peptide containing a tris-thiolate2 site engineered by d-cysteine ligands
92 c5u9tB_
not modelled
5.8
58
PDB header: de novo proteinChain: B: PDB Molecule: zn(ii)cl(coilser l16(dcy))3 2-;PDBTitle: the tris-thiolate zn(ii)s3cl binding site engineered by d-cysteine2 ligands in de novo three-stranded coiled coil environment
93 c3h5gA_
not modelled
5.8
58
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l16d-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
94 c3h5fA_
not modelled
5.8
58
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l16l-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
95 c3h5fC_
not modelled
5.8
58
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l16l-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
96 c3h5gC_
not modelled
5.8
58
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l16d-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
97 c5u9uB_
not modelled
5.8
58
PDB header: de novo proteinChain: B: PDB Molecule: apo-(coilser l16(dcy))3;PDBTitle: de novo three-stranded coiled coil peptide containing a tris-thiolate2 site engineered by d-cysteine ligands
98 c3h5gB_
not modelled
5.8
58
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l16d-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
99 c5u9tA_
not modelled
5.8
58
PDB header: de novo proteinChain: A: PDB Molecule: zn(ii)cl(coilser l16(dcy))3 2-;PDBTitle: the tris-thiolate zn(ii)s3cl binding site engineered by d-cysteine2 ligands in de novo three-stranded coiled coil environment