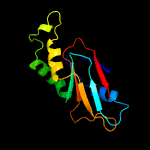
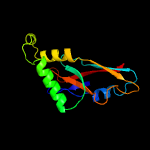
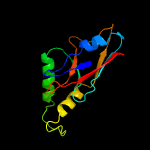
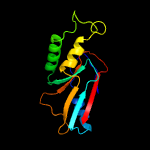
1 c2p6cB_
100.0
28
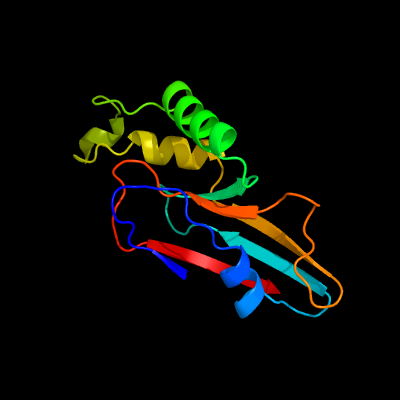
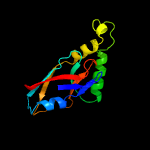
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: aq_2013 protein;PDBTitle: crystal structure of hypothetical protein aq_2013 from aquifex2 aeolicus vf5.
2 d1vpha_
100.0
29
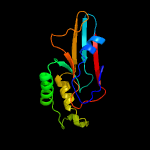
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like
3 d1vmja_
100.0
22
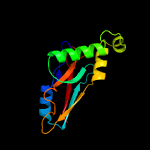
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like
4 c1ve0A_
100.0
28
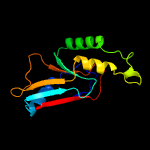
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein (st2072);PDBTitle: crystal structure of uncharacterized protein st2072 from sulfolobus2 tokodaii
5 c2p6hB_
100.0
32
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of hypothetical protein ape1520 from aeropyrum2 pernix k1
6 d1vmha_
100.0
26
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like
7 d1vmfa_
100.0
24
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like
8 c2cu5C_
100.0
28
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: conserved hypothetical protein tt1486;PDBTitle: crystal structure of the conserved hypothetical protein tt1486 from2 thermus thermophilus hb8
9 d1ojja_
38.1
11
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core
10 c2rfyB_
35.6
12
PDB header: hydrolaseChain: B: PDB Molecule: cellulose 1,4-beta-cellobiosidase;PDBTitle: crystal structure of cellobiohydrolase from melanocarpus albomyces2 complexed with cellobiose
11 c4zzpB_
34.8
17
PDB header: hydrolaseChain: B: PDB Molecule: cellulose 1,4-beta-cellobiosidase;PDBTitle: dictyostelium purpureum cellobiohydrolase cel7a apo structure
12 d1gpia_
34.8
15
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core
13 c2yg1A_
34.7
14
PDB header: hydrolaseChain: A: PDB Molecule: cellulose 1,4-beta-cellobiosidase;PDBTitle: apo structure of cellobiohydrolase 1 (cel7a) from heterobasidion2 annosum
14 d2v3ia1
33.8
14
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core
15 c4csiB_
33.4
17
PDB header: hydrolaseChain: B: PDB Molecule: cellulase;PDBTitle: crystal structure of the thermostable cellobiohydrolase2 cel7a from the fungus humicola grisea var. thermoidea.
16 d1q9ha_
31.3
12
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core
17 c2mdaB_
31.2
13
PDB header: oxidoreductaseChain: B: PDB Molecule: tyrosine 3-monooxygenase;PDBTitle: the solution structure of the regulatory domain of tyrosine2 hydroxylase
18 d3ovwa_
30.2
15
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core
19 c4hapB_
30.2
12
PDB header: hydrolaseChain: B: PDB Molecule: gh7 family protein;PDBTitle: crystal structure of a gh7 family cellobiohydrolase from limnoria2 quadripunctata in complex with cellobiose
20 d2qmwa2
29.4
7
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain
21 c4xnnA_
not modelled
28.7
15
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase chbi;PDBTitle: crystal structure of a gh7 family cellobiohydrolase from daphnia pulex
22 c3hi2D_
not modelled
25.2
28
PDB header: dna binding protein/toxinChain: D: PDB Molecule: motility quorum-sensing regulator mqsr;PDBTitle: structure of the n-terminal domain of the e. coli antitoxin mqsa2 (ygit/b3021) in complex with the e. coli toxin mqsr (ygiu/b3022)
23 d1eg1a_
not modelled
25.2
12
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolase family 7 catalytic core
24 c5o51A_
not modelled
19.3
25
PDB header: protein bindingChain: A: PDB Molecule: rho guanyl nucleotide exchange factor (rom2), putative;PDBTitle: afrom2 cnh domain
25 d1n8ia_
not modelled
19.2
21
Fold: TIM beta/alpha-barrelSuperfamily: Malate synthase GFamily: Malate synthase G
26 c3luyA_
not modelled
16.8
21
PDB header: isomeraseChain: A: PDB Molecule: probable chorismate mutase;PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
27 d1phza1
not modelled
15.8
7
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain
28 c3mwbA_
not modelled
15.7
7
PDB header: lyaseChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
29 d1i8fa_
not modelled
14.4
16
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
30 d2hq2a1
not modelled
13.1
20
Fold: Heme iron utilization protein-likeSuperfamily: Heme iron utilization protein-likeFamily: HemS/ChuS-like
31 c4lubA_
not modelled
12.2
7
PDB header: lyaseChain: A: PDB Molecule: putative prephenate dehydratase;PDBTitle: x-ray structure of prephenate dehydratase from streptococcus mutans
32 c5yvfD_
not modelled
12.0
20
PDB header: plant proteinChain: D: PDB Molecule: bfa1;PDBTitle: crystal structure of bfa1
33 c6c6lO_
not modelled
11.6
6
PDB header: membrane proteinChain: O: PDB Molecule: v-type proton atpase subunit f;PDBTitle: yeast vacuolar atpase vo in lipid nanodisc
34 d1cl1a_
not modelled
11.1
22
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like
35 c6cgvW_
not modelled
10.9
47
PDB header: virusChain: W: PDB Molecule: pre-protein vi;PDBTitle: revised crystal structure of human adenovirus
36 d1omwa2
not modelled
10.8
31
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
37 d1dq3a2
not modelled
10.5
26
Fold: dsRBD-likeSuperfamily: YcfA/nrd intein domainFamily: PI-Pfui intein middle domain
38 c3er0A_
not modelled
10.4
18
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 5a-2;PDBTitle: crystal structure of the full length eif5a from2 saccharomyces cerevisiae
39 c2qdrA_
not modelled
9.9
26
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative dioxygenase (npun_f5605) from nostoc2 punctiforme pcc 73102 at 2.60 a resolution
40 d2j0pa1
not modelled
9.9
14
Fold: Heme iron utilization protein-likeSuperfamily: Heme iron utilization protein-likeFamily: HemS/ChuS-like
41 d1ud2a1
not modelled
9.6
26
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
42 d2drpa2
not modelled
9.6
13
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
43 c3c9pA_
not modelled
9.5
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein sp1917;PDBTitle: crystal structure of uncharacterized protein sp1917
44 c6dgaA_
not modelled
8.3
17
PDB header: unknown functionChain: A: PDB Molecule: rpfr;PDBTitle: cronobacter turicensis rpfr quorum-sensing receptor rpff interaction2 domain
45 d1rutx4
not modelled
8.2
0
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain
46 c5dx5B_
not modelled
8.1
22
PDB header: lyaseChain: B: PDB Molecule: methionine gamma-lyase;PDBTitle: crystal structure of methionine gamma-lyase from clostridium2 sporogenes
47 c2gqnB_
not modelled
7.9
22
PDB header: lyaseChain: B: PDB Molecule: cystathionine beta-lyase;PDBTitle: cystathionine beta-lyase (cbl) from escherichia coli in complex with2 n-hydrazinocarbonylmethyl-2-nitro-benzamide
48 d1d8ca_
not modelled
7.7
22
Fold: TIM beta/alpha-barrelSuperfamily: Malate synthase GFamily: Malate synthase G
49 c2qmxB_
not modelled
7.2
7
PDB header: ligaseChain: B: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
50 c4mcjC_
not modelled
6.9
19
PDB header: transferaseChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative nucleoside deoxyribosyltransferase2 (bdi_0649) from parabacteroides distasonis atcc 8503 at 2.40 a3 resolution
51 c4ue0A_
not modelled
6.8
39
PDB header: viral proteinChain: A: PDB Molecule: fiber;PDBTitle: structure of the bovine atadenovirus type 4 fibre head protein
52 c2l57A_
not modelled
6.8
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of an uncharacterized thioredoin-like protein from2 clostridium perfringens
53 d1mwza_
not modelled
6.6
16
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain
54 c5dggB_
not modelled
6.3
38
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: central domain of uncharacterized lpg1148 protein from legionella2 pneumophila
55 d1xo8a_
not modelled
6.3
35
Fold: Immunoglobulin-like beta-sandwichSuperfamily: LEA14-likeFamily: LEA14-like
56 c2ogxB_
not modelled
6.3
13
PDB header: metal binding proteinChain: B: PDB Molecule: molybdenum storage protein subunit beta;PDBTitle: the crystal structure of the molybdenum storage protein from2 azotobacter vinelandii loaded with polyoxotungstates (wsto)
57 c3i38E_
not modelled
5.5
33
PDB header: chaperoneChain: E: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
58 c3i38C_
not modelled
5.5
33
PDB header: chaperoneChain: C: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
59 c3i38L_
not modelled
5.4
33
PDB header: chaperoneChain: L: PDB Molecule: putative chaperone dnaj;PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
60 c3jb9b_
not modelled
5.4
16
PDB header: rna binding protein/rnaChain: B: PDB Molecule: pre-mrna-splicing factor cwf10;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution