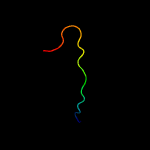| 1 |
|
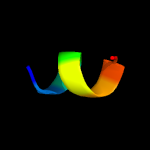
PDB 4pky chain E
Region: 3 - 25
Aligned: 23
Modelled: 23
Confidence: 11.7%
Identity: 39%
PDB header:transcription/cell cycle
Chain: E: PDB Molecule:transforming acidic coiled-coil-containing protein 3;
PDBTitle: arnt/hif transcription factor/coactivator complex
Phyre2
| 2 |
|
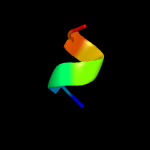
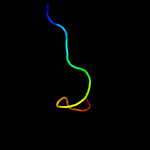
PDB 3thg chain A
Region: 35 - 65
Aligned: 29
Modelled: 31
Confidence: 11.1%
Identity: 28%
PDB header:protein binding
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase activase 1,
PDBTitle: crystal structure of the creosote rubisco activase c-domain
Phyre2
| 3 |
|
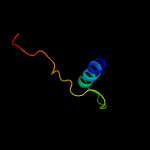
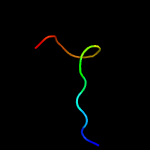
PDB 5u03 chain C
Region: 82 - 113
Aligned: 32
Modelled: 32
Confidence: 9.4%
Identity: 28%
PDB header:ligase, protein fibril
Chain: C: PDB Molecule:ctp synthase 1;
PDBTitle: cryo-em structure of the human ctp synthase filament
Phyre2
| 4 |
|
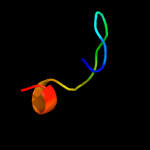
PDB 1l3a chain C
Region: 100 - 108
Aligned: 9
Modelled: 9
Confidence: 9.4%
Identity: 56%
Fold: ssDNA-binding transcriptional regulator domain
Superfamily: ssDNA-binding transcriptional regulator domain
Family: Plant transcriptional regulator PBF-2
Phyre2
| 5 |
|
PDB 2muq chain A
Region: 40 - 47
Aligned: 8
Modelled: 8
Confidence: 7.2%
Identity: 75%
PDB header:ubiquitin binding protein
Chain: A: PDB Molecule:fanconi anemia-associated protein of 20 kda;
PDBTitle: solution structure of the human faap20 ubz
Phyre2
| 6 |
|
PDB 3n1h chain A
Region: 100 - 107
Aligned: 8
Modelled: 8
Confidence: 7.0%
Identity: 50%
PDB header:dna binding protein
Chain: A: PDB Molecule:stwhy2;
PDBTitle: crystal structure of stwhy2
Phyre2
| 7 |
|
PDB 2mur chain A
Region: 40 - 47
Aligned: 8
Modelled: 8
Confidence: 6.6%
Identity: 75%
PDB header:protein binding
Chain: A: PDB Molecule:fanconi anemia-associated protein of 20 kda;
PDBTitle: solution structure of the human faap20 ubz-ubiquitin complex
Phyre2
| 8 |
|
PDB 1l3a chain A
Region: 100 - 107
Aligned: 8
Modelled: 8
Confidence: 6.5%
Identity: 63%
Fold: ssDNA-binding transcriptional regulator domain
Superfamily: ssDNA-binding transcriptional regulator domain
Family: Plant transcriptional regulator PBF-2
Phyre2
| 9 |
|
PDB 1l3a chain D
Region: 100 - 107
Aligned: 8
Modelled: 8
Confidence: 6.5%
Identity: 63%
PDB header:dna binding protein
Chain: D: PDB Molecule:p24: plant transcriptional regulator pbf-2;
PDBTitle: structure of the plant transcriptional regulator pbf-2
Phyre2
| 10 |
|
PDB 1p94 chain A
Region: 107 - 113
Aligned: 7
Modelled: 7
Confidence: 6.5%
Identity: 71%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: CopG-like
Phyre2
| 11 |
|
PDB 4xhi chain B
Region: 78 - 96
Aligned: 19
Modelled: 19
Confidence: 6.5%
Identity: 42%
PDB header:transcription
Chain: B: PDB Molecule:rna-dependent rna polymerase;
PDBTitle: crystal structure of native thosea asigna virus rna-dependent rna2 polymerase (rdrp) at 2.15 angstrom resolution
Phyre2
| 12 |
|
PDB 3dra chain B
Region: 75 - 89
Aligned: 15
Modelled: 15
Confidence: 6.4%
Identity: 40%
PDB header:transferase
Chain: B: PDB Molecule:geranylgeranyltransferase type i beta subunit;
PDBTitle: candida albicans protein geranylgeranyltransferase-i2 complexed with ggpp
Phyre2
| 13 |
|
PDB 5l85 chain B
Region: 38 - 72
Aligned: 28
Modelled: 35
Confidence: 6.2%
Identity: 32%
PDB header:signaling protein
Chain: B: PDB Molecule:nuclear fragile x mental retardation-interacting protein 1;
PDBTitle: solution structure of the complex between human znhit3 and nufip12 proteins
Phyre2
| 14 |
|
PDB 3j0c chain H
Region: 59 - 86
Aligned: 25
Modelled: 28
Confidence: 6.1%
Identity: 16%
PDB header:virus
Chain: H: PDB Molecule:e2 envelope glycoprotein;
PDBTitle: models of e1, e2 and cp of venezuelan equine encephalitis virus tc-832 strain restrained by a near atomic resolution cryo-em map
Phyre2
| 15 |
|
PDB 1tnu chain L
Region: 75 - 89
Aligned: 15
Modelled: 15
Confidence: 6.1%
Identity: 40%
PDB header:transferase
Chain: L: PDB Molecule:geranylgeranyl transferase type i beta subunit;
PDBTitle: rat protein geranylgeranyltransferase type-i complexed with2 a ggpp analog and a gcincckvl peptide derived from rhob
Phyre2
| 16 |
|
PDB 1n4q chain B
Region: 75 - 89
Aligned: 15
Modelled: 15
Confidence: 6.1%
Identity: 40%
Fold: alpha/alpha toroid
Superfamily: Terpenoid cyclases/Protein prenyltransferases
Family: Protein prenyltransferases
Phyre2
| 17 |
|
PDB 1kf6 chain A domain 1
Region: 63 - 80
Aligned: 18
Modelled: 18
Confidence: 6.1%
Identity: 39%
Fold: Spectrin repeat-like
Superfamily: Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domain
Family: Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domain
Phyre2
| 18 |
|
PDB 1dml chain H
Region: 112 - 124
Aligned: 13
Modelled: 13
Confidence: 5.9%
Identity: 62%
PDB header:dna binding protein/transferase
Chain: H: PDB Molecule:dna polymerase;
PDBTitle: crystal structure of herpes simplex ul42 bound to the c-terminus of2 hsv pol
Phyre2
| 19 |
|
PDB 1dml chain F
Region: 112 - 124
Aligned: 13
Modelled: 13
Confidence: 5.9%
Identity: 62%
PDB header:dna binding protein/transferase
Chain: F: PDB Molecule:dna polymerase;
PDBTitle: crystal structure of herpes simplex ul42 bound to the c-terminus of2 hsv pol
Phyre2
| 20 |
|
PDB 1dml chain B
Region: 112 - 124
Aligned: 13
Modelled: 13
Confidence: 5.8%
Identity: 62%
PDB header:dna binding protein/transferase
Chain: B: PDB Molecule:dna polymerase;
PDBTitle: crystal structure of herpes simplex ul42 bound to the c-terminus of2 hsv pol
Phyre2
| 21 |
|