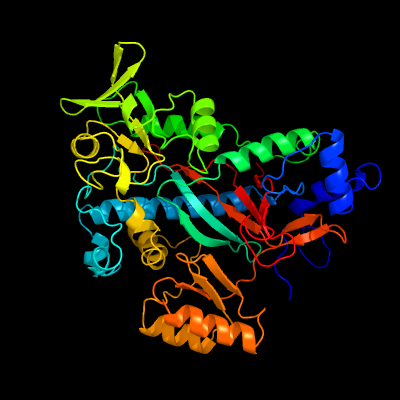
1 c3n6xA_
100.0
30
PDB header: ligaseChain: A: PDB Molecule: putative glutathionylspermidine synthase;PDBTitle: crystal structure of a putative glutathionylspermidine synthase2 (mfla_0391) from methylobacillus flagellatus kt at 2.35 a resolution
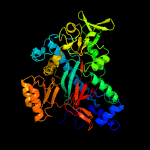
2 c2ioaA_
99.2
16
PDB header: ligase, hydrolaseChain: A: PDB Molecule: bifunctional glutathionylspermidinePDBTitle: e. coli bifunctional glutathionylspermidine2 synthetase/amidase incomplex with mg2+ and adp and3 phosphinate inhibitor
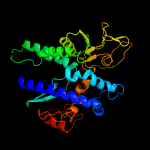
3 c2vpmB_
99.1
15
PDB header: ligaseChain: B: PDB Molecule: trypanothione synthetase;PDBTitle: trypanothione synthetase
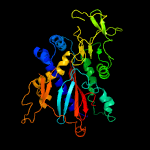
4 c1gshA_
96.5
15
PDB header: glutathione biosynthesis ligaseChain: A: PDB Molecule: glutathione biosynthetic ligase;PDBTitle: structure of escherichia coli glutathione synthetase at ph 7.5
5 d2io8a3
95.3
14
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: Glutathionylspermidine synthase ATP-binding domain-like
6 c4iwyA_
90.1
13
PDB header: ligaseChain: A: PDB Molecule: ribosomal protein s6 modification protein;PDBTitle: semet-substituted rimk structure
7 c1m0tB_
86.2
14
PDB header: ligaseChain: B: PDB Molecule: glutathione synthetase;PDBTitle: yeast glutathione synthase
8 c1i7nA_
86.1
13
PDB header: neuropeptideChain: A: PDB Molecule: synapsin ii;PDBTitle: crystal structure analysis of the c domain of synapsin ii2 from rat brain
9 c5i47A_
84.7
10
PDB header: biosynthetic proteinChain: A: PDB Molecule: rimk domain protein atp-grasp;PDBTitle: crystal structure of rimk domain protein atp-grasp from sphaerobacter2 thermophilus dsm 20745
10 c3vpbC_
80.1
21
PDB header: ligaseChain: C: PDB Molecule: putative acetylornithine deacetylase;PDBTitle: argx from sulfolobus tokodaii complexed with2 lysw/glu/adp/mg/zn/sulfate
11 c1kjjA_
79.4
17
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase 2;PDBTitle: crystal structure of glycniamide ribonucleotide transformylase in2 complex with mg-atp-gamma-s
12 c3kalB_
77.4
11
PDB header: ligaseChain: B: PDB Molecule: homoglutathione synthetase;PDBTitle: structure of homoglutathione synthetase from glycine max in2 closed conformation with homoglutathione, adp, a sulfate3 ion, and three magnesium ions bound
13 c1pk8D_
76.4
13
PDB header: membrane proteinChain: D: PDB Molecule: rat synapsin i;PDBTitle: crystal structure of rat synapsin i c domain complexed to2 ca.atp
14 c3wvqA_
75.8
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: pgm1;PDBTitle: structure of atp grasp protein
15 c3tinA_
75.1
22
PDB header: ligaseChain: A: PDB Molecule: ttl protein;PDBTitle: tubulin tyrosine ligase
16 c5ig9H_
73.9
17
PDB header: ligaseChain: H: PDB Molecule: atp grasp ligase;PDBTitle: crystal structure of macrocyclase mdnc bound with precursor peptide2 mdna from microcystis aeruginosa mrc
17 d1pk8a2
71.9
14
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: Synapsin C-terminal domain
18 c5ig8A_
71.5
14
PDB header: ligaseChain: A: PDB Molecule: atp grasp ligase;PDBTitle: crystal structure of macrocyclase mdnb from microcystis aeruginosa mrc
19 c1ehiB_
71.3
11
PDB header: ligaseChain: B: PDB Molecule: d-alanine:d-lactate ligase;PDBTitle: d-alanine:d-lactate ligase (lmddl2) of vancomycin-resistant2 leuconostoc mesenteroides
20 c2p0aA_
70.4
14
PDB header: neuropeptideChain: A: PDB Molecule: synapsin-3;PDBTitle: the crystal structure of human synapsin iii (syn3) in complex with2 amppnp
21 d1gsaa2
not modelled
69.3
12
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: ATP-binding domain of peptide synthetases
22 d1i7na2
not modelled
68.7
14
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: Synapsin C-terminal domain
23 c2qb5B_
not modelled
67.7
18
PDB header: transferaseChain: B: PDB Molecule: inositol-tetrakisphosphate 1-kinase;PDBTitle: crystal structure of human inositol 1,3,4-trisphosphate 5/6-kinase2 (itpk1) in complex with adp and mn2+
24 d1kjqa3
not modelled
67.0
14
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like
25 c3i12A_
not modelled
66.5
21
PDB header: ligaseChain: A: PDB Molecule: d-alanine-d-alanine ligase a;PDBTitle: the crystal structure of the d-alanyl-alanine synthetase a from2 salmonella enterica subsp. enterica serovar typhimurium str. lt2
26 c3lp8A_
not modelled
65.5
9
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylamine-glycine ligase;PDBTitle: crystal structure of phosphoribosylamine-glycine ligase from2 ehrlichia chaffeensis
27 c5oetB_
not modelled
64.9
14
PDB header: ligaseChain: B: PDB Molecule: glutathione synthetase-like effector 30 (gpa-gss30-apo);PDBTitle: the structure of a glutathione synthetase like-effector (gss30) from2 globodera pallida in apoform.
28 c5oevB_
not modelled
60.7
14
PDB header: transferaseChain: B: PDB Molecule: glutathione synthetase-like effector 22 (gpa-gss22-apo);PDBTitle: the structure of a glutathione synthetase like-effector (gss22) from2 globodera pallida in apoform.
29 c1e4eB_
not modelled
59.7
23
PDB header: ligaseChain: B: PDB Molecule: vancomycin/teicoplanin a-type resistance protein vana;PDBTitle: d-alanyl-d-lacate ligase
30 d1iowa2
not modelled
57.5
20
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: ATP-binding domain of peptide synthetases
31 c4fu0B_
not modelled
56.8
15
PDB header: ligaseChain: B: PDB Molecule: d-alanine--d-alanine ligase 7;PDBTitle: crystal structure of vang d-ala:d-ser ligase from enterococcus2 faecalis
32 c3e5nA_
not modelled
53.6
21
PDB header: ligaseChain: A: PDB Molecule: d-alanine-d-alanine ligase a;PDBTitle: crystal strucutre of d-alanine-d-alanine ligase from2 xanthomonas oryzae pv. oryzae kacc10331
33 c4iijF_
not modelled
53.4
16
PDB header: cell cycleChain: F: PDB Molecule: tubulin tyrosine ligase, ttl;PDBTitle: crystal structure of tubulin-stathmin-ttl-apo complex
34 c3tqtB_
not modelled
52.7
13
PDB header: ligaseChain: B: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: structure of the d-alanine-d-alanine ligase from coxiella burnetii
35 c4wd3B_
not modelled
52.7
9
PDB header: ligaseChain: B: PDB Molecule: l-amino acid ligase;PDBTitle: crystal structure of an l-amino acid ligase riza
36 c6dgiA_
not modelled
51.6
17
PDB header: ligaseChain: A: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: the crystal structure of d-alanyl-alanine synthetase a from vibrio2 cholerae o1 biovar eltor str. n16961
37 c5cviB_
not modelled
50.3
12
PDB header: transcription regulatorChain: B: PDB Molecule: slor;PDBTitle: structure of the manganese regulator slor
38 c1w96B_
not modelled
48.8
16
PDB header: ligaseChain: B: PDB Molecule: acetyl-coenzyme a carboxylase;PDBTitle: crystal structure of biotin carboxylase domain of acetyl-2 coenzyme a carboxylase from saccharomyces cerevisiae in3 complex with soraphen a
39 c2yyaB_
not modelled
47.0
10
PDB header: ligaseChain: B: PDB Molecule: phosphoribosylamine--glycine ligase;PDBTitle: crystal structure of gar synthetase from aquifex aeolicus
40 c2dlnA_
not modelled
46.3
23
PDB header: ligase(peptidoglycan synthesis)Chain: A: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: vancomycin resistance: structure of d-alanine:d-alanine ligase at 2.32 angstroms resolution
41 d1e4ea2
not modelled
45.7
18
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: ATP-binding domain of peptide synthetases
42 d2gxfa1
not modelled
45.1
12
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: YybH-like
43 d1a9xa5
not modelled
44.6
13
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like
44 d1omha_
not modelled
42.9
20
Fold: Origin of replication-binding domain, RBD-likeSuperfamily: Origin of replication-binding domain, RBD-likeFamily: Relaxase domain
45 c3soyA_
not modelled
41.1
10
PDB header: membrane proteinChain: A: PDB Molecule: ntf2-like superfamily protein;PDBTitle: nuclear transport factor 2 (ntf2-like) superfamily protein from2 salmonella enterica subsp. enterica serovar typhimurium str. lt2
46 c2ip4A_
not modelled
40.1
15
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylamine--glycine ligase;PDBTitle: crystal structure of glycinamide ribonucleotide synthetase from2 thermus thermophilus hb8
47 c5kciA_
not modelled
39.9
17
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein ypl067c;PDBTitle: crystal structure of htc1
48 c2xd4A_
not modelled
39.5
12
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylamine--glycine ligase;PDBTitle: nucleotide-bound structures of bacillus subtilis glycinamide2 ribonucleotide synthetase
49 c4mamB_
not modelled
38.5
10
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: the crystal structure of phosphoribosylaminoimidazole carboxylase2 atpase subunit of francisella tularensis subsp. tularensis schu s4 in3 complex with an adp analog, amp-cp
50 c2i80B_
not modelled
38.0
14
PDB header: ligaseChain: B: PDB Molecule: d-alanine-d-alanine ligase;PDBTitle: allosteric inhibition of staphylococcus aureus d-alanine:d-alanine2 ligase revealed by crystallographic studies
51 c3se7A_
not modelled
37.2
35
PDB header: ligaseChain: A: PDB Molecule: vana;PDBTitle: ancient vana
52 d1uc8a2
not modelled
37.0
13
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: Lysine biosynthesis enzyme LysX ATP-binding domain
53 c1z2pX_
not modelled
36.8
21
PDB header: transferaseChain: X: PDB Molecule: inositol 1,3,4-trisphosphate 5/6-kinase;PDBTitle: inositol 1,3,4-trisphosphate 5/6-kinase in complex with mg2+/amp-2 pcp/ins(1,3,4)p3
54 c5k2mG_
not modelled
35.6
21
PDB header: biosynthetic proteinChain: G: PDB Molecule: rimk-related lysine biosynthesis protein;PDBTitle: bifunctional lysx/argx from thermococcus kodakarensis with lysw-gamma-2 aaa
55 c6o5cB_
not modelled
35.2
15
PDB header: dna binding proteinChain: B: PDB Molecule: putative metal-dependent transcriptional regulator;PDBTitle: x-ray crystal structure of metal-dependent transcriptional regulator2 mtsr
56 c1vkzA_
not modelled
33.9
12
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylamine--glycine ligase;PDBTitle: crystal structure of phosphoribosylamine--glycine ligase (tm1250) from2 thermotoga maritima at 2.30 a resolution
57 c3oetF_
not modelled
33.8
19
PDB header: oxidoreductaseChain: F: PDB Molecule: erythronate-4-phosphate dehydrogenase;PDBTitle: d-erythronate-4-phosphate dehydrogenase complexed with nad
58 d1gsoa3
not modelled
33.5
14
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like
59 d1ewqa4
not modelled
31.9
22
Fold: MutS N-terminal domain-likeSuperfamily: DNA repair protein MutS, domain IFamily: DNA repair protein MutS, domain I
60 c3gidB_
not modelled
31.1
16
PDB header: ligaseChain: B: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: the biotin carboxylase (bc) domain of human acetyl-coa carboxylase 22 (acc2) in complex with soraphen a
61 c5oevD_
not modelled
31.0
13
PDB header: transferaseChain: D: PDB Molecule: glutathione synthetase-like effector 22 (gpa-gss22-apo);PDBTitle: the structure of a glutathione synthetase like-effector (gss22) from2 globodera pallida in apoform.
62 c3ln6A_
not modelled
29.9
9
PDB header: ligaseChain: A: PDB Molecule: glutathione biosynthesis bifunctional protein gshab;PDBTitle: crystal structure of a bifunctional glutathione synthetase from2 streptococcus agalactiae
63 c4egqD_
not modelled
28.3
14
PDB header: ligaseChain: D: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: crystal structure of d-alanine-d-alanine ligase b from burkholderia2 pseudomallei
64 d2r85a2
not modelled
28.1
18
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: PurP ATP-binding domain-like
65 c6jdxC_
not modelled
27.3
16
PDB header: hydrolase/hydrolase inhibitorChain: C: PDB Molecule: crispr-associated endonuclease cas9;PDBTitle: crystal structure of acriic2 dimer in complex with partial nme1cas92 preprocessed with protease alpha-chymotrypsin
66 d3c0na2
not modelled
27.2
16
Fold: Aerolisin/ETX pore-forming domainSuperfamily: Aerolisin/ETX pore-forming domainFamily: (Pro)aerolysin, pore-forming lobe
67 c2hjwA_
not modelled
26.4
17
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: crystal structure of the bc domain of acc2
68 c4qdfA_
not modelled
24.6
16
PDB header: oxidoreductase/oxidoreductase inhibitorChain: A: PDB Molecule: 3-ketosteroid 9alpha-hydroxylase oxygenase;PDBTitle: crystal structure of apo ksha5 and ksha1 in complex with 1,4-30q-coa2 from r. rhodochrous
69 c3c8iA_
not modelled
23.8
27
PDB header: membrane proteinChain: A: PDB Molecule: putative membrane protein;PDBTitle: crystal structure of a putative membrane protein from corynebacterium2 diphtheriae
70 c3h75A_
not modelled
23.1
37
PDB header: sugar binding proteinChain: A: PDB Molecule: periplasmic sugar-binding domain protein;PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
71 c3orqA_
not modelled
22.7
10
PDB header: ligase,biosynthetic proteinChain: A: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide synthetase;PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
72 c2zdqA_
not modelled
22.0
23
PDB header: ligaseChain: A: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: crystal structure of d-alanine:d-alanine ligase with atp2 and d-alanine:d-alanine from thermus thermophius hb8
73 c5dt9A_
not modelled
21.7
20
PDB header: oxidoreductaseChain: A: PDB Molecule: erythronate-4-phosphate dehydrogenase;PDBTitle: crystal structure of a putative d-erythronate-4-phosphate2 dehydrogenase from vibrio cholerae
74 c1wtaA_
not modelled
21.5
20
PDB header: transferaseChain: A: PDB Molecule: 5'-methylthioadenosine phosphorylase;PDBTitle: crystal structure of 5'-deoxy-5'-methylthioadenosine from aeropyrum2 pernix (r32 form)
75 c4njmA_
not modelled
21.2
21
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase, putative;PDBTitle: crystal structure of phosphoglycerate bound 3-phosphoglycerate2 dehydrogenase in entamoeba histolytica
76 c6fanC_
not modelled
21.0
23
PDB header: metal binding proteinChain: C: PDB Molecule: coot;PDBTitle: crystal structure of putative coot from carboxydothermus2 hydrogenoformans
77 d2r7ka2
not modelled
20.3
8
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: PurP ATP-binding domain-like
78 c2o4cB_
not modelled
20.0
21
PDB header: oxidoreductaseChain: B: PDB Molecule: erythronate-4-phosphate dehydrogenase;PDBTitle: crystal structure of d-erythronate-4-phosphate dehydrogenase complexed2 with nad
79 c3ll4B_
not modelled
19.7
7
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein ykr043c;PDBTitle: structure of the h13a mutant of ykr043c in complex with fructose-1,6-2 bisphosphate
80 c5diyB_
not modelled
19.4
15
PDB header: hydrolaseChain: B: PDB Molecule: hyaluronidase;PDBTitle: thermobaculum terrenum o-glcnac hydrolase mutant - d120n
81 c3dktD_
not modelled
19.3
13
PDB header: structural protein/virus like particleChain: D: PDB Molecule: maritimacin;PDBTitle: crystal structure of thermotoga maritima encapsulin
82 c1ik6A_
not modelled
18.6
22
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate dehydrogenase;PDBTitle: 3d structure of the e1beta subunit of pyruvate2 dehydrogenase from the archeon pyrobaculum aerophilum
83 c2e5qA_
not modelled
18.5
17
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 19;PDBTitle: solution structure of the tudor domain of phd finger2 protein 19, isoform b [homo sapiens]
84 c3hruA_
not modelled
18.1
15
PDB header: transcriptionChain: A: PDB Molecule: metalloregulator scar;PDBTitle: crystal structure of scar with bound zn2+
85 c4prkB_
not modelled
17.7
20
PDB header: oxidoreductaseChain: B: PDB Molecule: 4-phosphoerythronate dehydrogenase;PDBTitle: crystal structure of d-lactate dehydrogenase (d-ldh) from2 lactobacillus jensenii
86 d2dlda2
not modelled
17.7
10
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain
87 c3ecjC_
not modelled
17.3
11
PDB header: oxidoreductaseChain: C: PDB Molecule: protein (homoprotocatechuate 2,3-dioxygenase);PDBTitle: structure of e323l mutant of homoprotocatechuate 2,3-dioxygenase from2 brevibacterium fuscum at 1.65a resolution
88 c4p16A_
not modelled
17.2
20
PDB header: hydrolaseChain: A: PDB Molecule: orf1a;PDBTitle: crystal structure of the papain-like protease of middle-east2 respiratory syndrome coronavirus
89 c4yakD_
not modelled
17.1
7
PDB header: ligaseChain: D: PDB Molecule: beta subunit of acyl-coa synthetase (ndp forming);PDBTitle: ca. korarchaeum cryptofilum dinucleotide forming acetyl-coenzyme a2 synthetase 1 in complex with coenzyme a, acetyl-coenzyme a and with3 phosphorylated phosphohistidine segment (site i orientation)
90 c2lc0A_
not modelled
17.0
24
PDB header: protein bindingChain: A: PDB Molecule: putative uncharacterized protein tb39.8;PDBTitle: rv0020c_nter structure
91 c3wwyA_
not modelled
17.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: d-lactate dehydrogenase;PDBTitle: the crystal structure of d-lactate dehydrogenase from fusobacterium2 nucleatum subsp. nucleatum
92 c3bb9D_
not modelled
16.9
19
PDB header: unknown functionChain: D: PDB Molecule: putative orphan protein;PDBTitle: crystal structure of a putative ketosteroid isomerase (sfri_1973) from2 shewanella frigidimarina ncimb 400 at 1.80 a resolution
93 c3bg5C_
not modelled
16.6
20
PDB header: ligaseChain: C: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of staphylococcus aureus pyruvate carboxylase
94 c2e5pA_
not modelled
16.2
21
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 1 (phf1 protein)
95 c3wwzB_
not modelled
16.1
18
PDB header: oxidoreductaseChain: B: PDB Molecule: d-lactate dehydrogenase (fermentative);PDBTitle: the crystal structure of d-lactate dehydrogenase from pseudomonas2 aeruginosa
96 c2hwgA_
not modelled
16.1
20
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of phosphorylated enzyme i of the phosphoenolpyruvate:sugar2 phosphotransferase system
97 c4qdfB_
not modelled
16.0
6
PDB header: oxidoreductase/oxidoreductase inhibitorChain: B: PDB Molecule: 3-ketosteroid 9alpha-hydroxylase oxygenase;PDBTitle: crystal structure of apo ksha5 and ksha1 in complex with 1,4-30q-coa2 from r. rhodochrous
98 c3r23B_
not modelled
15.9
19
PDB header: ligaseChain: B: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: crystal structure of d-alanine--d-alanine ligase from bacillus2 anthracis
99 c2pvpB_
not modelled
15.7
13
PDB header: ligaseChain: B: PDB Molecule: d-alanine-d-alanine ligase;PDBTitle: crystal structure of d-alanine-d-alanine ligase from helicobacter2 pylori