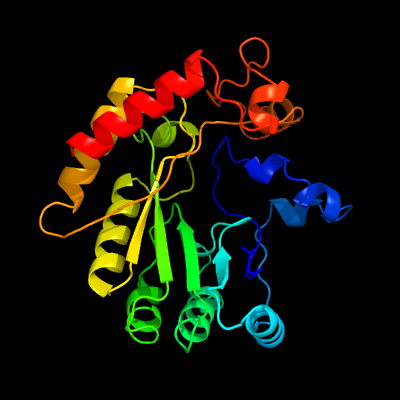
| 1 | c4fheA_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:spore photoproduct lyase;
PDBTitle: spore photoproduct lyase c140a mutant
|
|
|
|
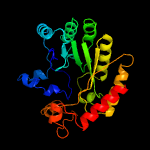
| 2 | c4jc0B_
|
|
 |
100.0 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:ribosomal protein s12 methylthiotransferase rimo;
PDBTitle: crystal structure of thermotoga maritima holo rimo in complex with2 pentasulfide, northeast structural genomics consortium target vr77
|
|
|
|
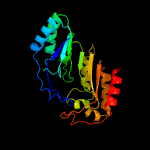
| 3 | c6fd2B_
|
|
 |
99.9 |
19 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:putative apramycin biosynthetic oxidoreductase 4;
PDBTitle: radical sam 1,2-diol dehydratase aprd4 in complex with its substrate2 paromamine
|
|
|
|
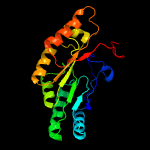
| 4 | d1olta_
|
|
 |
99.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Oxygen-independent coproporphyrinogen III oxidase HemN |
|
|
|
| 5 | c5l7jA_
|
|
 |
99.9 |
16 |
PDB header:translation
Chain: A: PDB Molecule:elp3 family;
PDBTitle: crystal structure of elp3 from dehalococcoides mccartyi
|
|
|
|
| 6 | c5ul4A_
|
|
 |
99.9 |
12 |
PDB header:metal binding protein
Chain: A: PDB Molecule:oxsb protein;
PDBTitle: structure of cobalamin-dependent s-adenosylmethionine radical enzyme2 oxsb with aqua-cobalamin and s-adenosylmethionine bound
|
|
|
|
| 7 | c6qk7C_
|
|
 |
99.9 |
15 |
PDB header:translation
Chain: C: PDB Molecule:elongator complex protein 3;
PDBTitle: elongator catalytic subcomplex elp123 lobe
|
|
|
|
| 8 | c3cixA_
|
|
 |
99.9 |
17 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
|
|
|
| 9 | c2qgqF_
|
|
 |
99.9 |
10 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:protein tm_1862;
PDBTitle: crystal structure of tm_1862 from thermotoga maritima. northeast2 structural genomics consortium target vr77
|
|
|
|
| 10 | c3t7vA_
|
|
 |
99.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:methylornithine synthase pylb;
PDBTitle: crystal structure of methylornithine synthase (pylb)
|
|
|
|
| 11 | c5v1tA_
|
|
 |
99.8 |
17 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam;
PDBTitle: crystal structure of streptococcus suis suib bound to precursor2 peptide suia
|
|
|
|
| 12 | c5exkG_
|
|
 |
99.8 |
19 |
PDB header:transferase
Chain: G: PDB Molecule:lipoyl synthase;
PDBTitle: crystal structure of m. tuberculosis lipoyl synthase with 6-2 thiooctanoyl peptide intermediate
|
|
|
|
| 13 | c1r30A_
|
|
 |
99.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:biotin synthase;
PDBTitle: the crystal structure of biotin synthase, an s-adenosylmethionine-2 dependent radical enzyme
|
|
|
|
| 14 | d1r30a_
|
|
 |
99.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Biotin synthase |
|
|
|
| 15 | c6iazA_
|
|
 |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:histone acetyltransferase, elp3 family;
PDBTitle: the archaeal methanocaldococcus infernus elp3 with n-terminus deletion2 (1-46)
|
|
|
|
| 16 | c3rfaA_
|
|
 |
99.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribosomal rna large subunit methyltransferase n;
PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
|
|
|
|
| 17 | c3rfaB_
|
|
 |
99.7 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ribosomal rna large subunit methyltransferase n;
PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
|
|
|
|
| 18 | c6fz6B_
|
|
 |
99.7 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:probable dual-specificity rna methyltransferase rlmn;
PDBTitle: crystal structure of a radical sam methyltransferase from2 sphaerobacter thermophilus
|
|
|
|
| 19 | d1tv8a_
|
|
 |
99.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:MoCo biosynthesis proteins |
|
|
|
| 20 | c2yx0A_
|
|
 |
99.7 |
19 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam enzyme;
PDBTitle: crystal structure of p. horikoshii tyw1
|
|
|
|
| 21 | c6efnA_ |
|
not modelled |
99.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sporulation killing factor maturation protein skfb;
PDBTitle: structure of a ripp maturase, skfb
|
|
|
| 22 | c4u0pB_ |
|
not modelled |
99.7 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:lipoyl synthase 2;
PDBTitle: the crystal structure of lipoyl synthase in complex with s-adenosyl2 homocysteine
|
|
|
| 23 | c3c8fA_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate formate-lyase 1-activating enzyme;
PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with partially2 disordered adomet
|
|
|
| 24 | c4wcxC_ |
|
not modelled |
99.5 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:biotin and thiamin synthesis associated;
PDBTitle: crystal structure of hydg: a maturase of the [fefe]-hydrogenase
|
|
|
| 25 | c4k39A_ |
|
not modelled |
99.5 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:anaerobic sulfatase-maturating enzyme;
PDBTitle: native ansmecpe with bound adomet and cp18cys peptide
|
|
|
| 26 | c2a5hC_ |
|
not modelled |
99.5 |
14 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
|
|
| 27 | c5vslB_ |
|
not modelled |
99.5 |
12 |
PDB header:antiviral protein
Chain: B: PDB Molecule:radical s-adenosyl methionine domain-containing protein 2;
PDBTitle: crystal structure of viperin with bound [4fe-4s] cluster and s-2 adenosylhomocysteine (sah)
|
|
|
| 28 | c5wggA_ |
|
not modelled |
99.5 |
10 |
PDB header:peptide binding protein
Chain: A: PDB Molecule:radical sam domain protein;
PDBTitle: structural insights into thioether bond formation in the biosynthesis2 of sactipeptides
|
|
|
| 29 | c4rtbA_ |
|
not modelled |
99.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:hydg protein;
PDBTitle: x-ray structure of the fefe-hydrogenase maturase hydg from2 carboxydothermus hydrogenoformans
|
|
|
| 30 | c6b4cH_ |
|
not modelled |
99.4 |
10 |
PDB header:antiviral protein
Chain: H: PDB Molecule:viperin;
PDBTitle: structure of viperin from trichoderma virens
|
|
|
| 31 | c5th5C_ |
|
not modelled |
99.4 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from bacillus subtilis with 6-carboxypterin-2 5'-deoxyadenosyl ester bound
|
|
|
| 32 | c4r33A_ |
|
not modelled |
99.1 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:nosl;
PDBTitle: x-ray structure of the tryptophan lyase nosl with tryptophan and s-2 adenosyl-l-homocysteine bound
|
|
|
| 33 | c6nhlB_ |
|
not modelled |
99.0 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from escherichia coli
|
|
|
| 34 | c4m7tA_ |
|
not modelled |
98.9 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:btrn;
PDBTitle: crystal structure of btrn in complex with adomet and 2-doia
|
|
|
| 35 | c3canA_ |
|
not modelled |
98.8 |
11 |
PDB header:lyase activator
Chain: A: PDB Molecule:pyruvate-formate lyase-activating enzyme;
PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
|
|
|
| 36 | c2z2uA_ |
|
not modelled |
98.7 |
14 |
PDB header:metal binding protein
Chain: A: PDB Molecule:upf0026 protein mj0257;
PDBTitle: crystal structure of archaeal tyw1
|
|
|
| 37 | c4njkA_ |
|
not modelled |
98.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from burkholderia multivorans in complex2 with adomet, 7-carboxy-7-deazaguanine, and mg2+
|
|
|
| 38 | c6c8vA_ |
|
not modelled |
98.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme pqq synthesis protein e;
PDBTitle: x-ray structure of pqqe from methylobacterium extorquens
|
|
|
| 39 | c5kzmA_ |
|
not modelled |
93.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
|
|
|
| 40 | c2y5sA_ |
|
not modelled |
92.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
|
|
| 41 | c3vndD_ |
|
not modelled |
92.1 |
24 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
|
|
|
| 42 | d1qopa_ |
|
not modelled |
91.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 43 | c5tchG_ |
|
not modelled |
90.8 |
19 |
PDB header:lyase
Chain: G: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
|
|
|
| 44 | d2p10a1 |
|
not modelled |
90.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Mll9387-like |
|
|
| 45 | c6omzA_ |
|
not modelled |
89.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from mycobacterium2 smegmatis with bound 6-hydroxymethylpterin-monophosphate
|
|
|
| 46 | c5k9xA_ |
|
not modelled |
88.1 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha chain from legionella2 pneumophila subsp. pneumophila
|
|
|
| 47 | c2ekcA_ |
|
not modelled |
87.7 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
|
|
| 48 | c3lmzA_ |
|
not modelled |
87.3 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase. (yp_001305105.1) from2 parabacteroides distasonis atcc 8503 at 1.44 a resolution
|
|
|
| 49 | c3p6lA_ |
|
not modelled |
86.6 |
8 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_1903)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
|
|
|
| 50 | c3navB_ |
|
not modelled |
86.5 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 51 | c2dzaA_ |
|
not modelled |
86.5 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
|
|
| 52 | d1x7fa2 |
|
not modelled |
86.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Outer surface protein, N-terminal domain |
|
|
| 53 | d1bxba_ |
|
not modelled |
85.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 54 | c5ey5A_ |
|
not modelled |
85.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:lbcats-a;
PDBTitle: lbcats
|
|
|
| 55 | c2zdsB_ |
|
not modelled |
85.2 |
10 |
PDB header:dna binding protein
Chain: B: PDB Molecule:putative dna-binding protein;
PDBTitle: crystal structure of sco6571 from streptomyces coelicolor a3(2)
|
|
|
| 56 | c3bleA_ |
|
not modelled |
84.6 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in complexed with2 malonate
|
|
|
| 57 | d1l6wa_ |
|
not modelled |
82.6 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 58 | c3rmjB_ |
|
not modelled |
82.4 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of truncated alpha-isopropylmalate synthase from2 neisseria meningitidis
|
|
|
| 59 | c5ks8D_ |
|
not modelled |
82.3 |
10 |
PDB header:ligase
Chain: D: PDB Molecule:pyruvate carboxylase subunit beta;
PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
|
|
|
| 60 | d1j0ha3 |
|
not modelled |
81.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 61 | c4ovxA_ |
|
not modelled |
81.6 |
7 |
PDB header:isomerase
Chain: A: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structure of xylose isomerase domain protein from planctomyces2 limnophilus dsm 3776
|
|
|
| 62 | c2z1kA_ |
|
not modelled |
81.5 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:(neo)pullulanase;
PDBTitle: crystal structure of ttha1563 from thermus thermophilus hb8
|
|
|
| 63 | c6e1jB_ |
|
not modelled |
81.3 |
13 |
PDB header:plant protein
Chain: B: PDB Molecule:2-isopropylmalate synthase, a genome specific 1;
PDBTitle: crystal structure of methylthioalkylmalate synthase (bjumam1.1) from2 brassica juncea
|
|
|
| 64 | c3s1vD_ |
|
not modelled |
80.9 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:probable transaldolase;
PDBTitle: transaldolase from thermoplasma acidophilum in complex with d-fructose2 6-phosphate schiff-base intermediate
|
|
|
| 65 | c1x7fA_ |
|
not modelled |
80.7 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:outer surface protein;
PDBTitle: crystal structure of an uncharacterized b. cereus protein
|
|
|
| 66 | d1wx0a1 |
|
not modelled |
80.6 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 67 | d1ea9c3 |
|
not modelled |
80.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 68 | c3vm7A_ |
|
not modelled |
80.1 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: structure of an alpha-amylase from malbranchea cinnamomea
|
|
|
| 69 | d1qt1a_ |
|
not modelled |
80.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 70 | c3ewbX_ |
|
not modelled |
79.8 |
13 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-isopropylmalate2 synthase from listeria monocytogenes
|
|
|
| 71 | c3cqkB_ |
|
not modelled |
79.5 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
|
|
| 72 | c5ud6B_ |
|
not modelled |
79.5 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dhdps from cyanidioschyzon merolae with lysine2 bound
|
|
|
| 73 | c1ydnA_ |
|
not modelled |
79.4 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
|
|
| 74 | c2bdqA_ |
|
not modelled |
79.0 |
12 |
PDB header:metal transport
Chain: A: PDB Molecule:copper homeostasis protein cutc;
PDBTitle: crystal structure of the putative copper homeostasis protein cutc from2 streptococcus agalactiae, northeast strucural genomics target sar15.
|
|
|
| 75 | d1pkla2 |
|
not modelled |
78.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
|
|
| 76 | c2ftpA_ |
|
not modelled |
78.7 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
|
|
| 77 | d1ajza_ |
|
not modelled |
78.4 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
| 78 | c5vrvA_ |
|
not modelled |
78.3 |
16 |
PDB header:hydrolase,oxidoreductase
Chain: A: PDB Molecule:protein regulated by acid ph;
PDBTitle: 2.05 angstrom resolution crystal structure of c-terminal domain2 (duf2156) of putative lysylphosphatidylglycerol synthetase from3 agrobacterium fabrum.
|
|
|
| 79 | d2q02a1 |
|
not modelled |
77.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
|
|
| 80 | d1muma_ |
|
not modelled |
77.6 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 81 | d1vpxa_ |
|
not modelled |
77.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 82 | c1ea9D_ |
|
not modelled |
77.2 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:cyclomaltodextrinase;
PDBTitle: cyclomaltodextrinase
|
|
|
| 83 | d1ujqa_ |
|
not modelled |
76.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 84 | d1yx1a1 |
|
not modelled |
76.8 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:KguE-like |
|
|
| 85 | c3qbuD_ |
|
not modelled |
76.5 |
12 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of putative peptidoglycan deactelyase (hp0310) from2 helicobacter pylori
|
|
|
| 86 | d2aaaa2 |
|
not modelled |
76.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 87 | c3khdC_ |
|
not modelled |
76.3 |
8 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pff1300w.
|
|
|
| 88 | c3thaB_ |
|
not modelled |
76.1 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
|
|
|
| 89 | c3bdkB_ |
|
not modelled |
74.6 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
|
|
| 90 | c3kwsB_ |
|
not modelled |
74.2 |
10 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
|
|
| 91 | c3d0cB_ |
|
not modelled |
73.1 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
|
|
|
| 92 | d1a3xa2 |
|
not modelled |
72.8 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
|
|
| 93 | c3dx5A_ |
|
not modelled |
72.7 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
|
|
| 94 | d1gvia3 |
|
not modelled |
72.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 95 | c3r8rJ_ |
|
not modelled |
72.5 |
22 |
PDB header:transferase
Chain: J: PDB Molecule:transaldolase;
PDBTitle: transaldolase from bacillus subtilis
|
|
|
| 96 | c2p10D_ |
|
not modelled |
72.1 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
|
|
| 97 | c5afdA_ |
|
not modelled |
71.9 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: native structure of n-acetylneuramininate lyase (sialic acid aldolase)2 from aliivibrio salmonicida
|
|
|
| 98 | c4k3zA_ |
|
not modelled |
71.7 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-erythrulose 4-phosphate dehydrogenase;
PDBTitle: crystal structure of d-erythrulose 4-phosphate dehydrogenase from2 brucella melitensis, solved by iodide sad
|
|
|
| 99 | d1djqa1 |
|
not modelled |
71.6 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 100 | d1twda_ |
|
not modelled |
71.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:CutC-like
Family:CutC-like |
|
|
| 101 | c3c52B_ |
|
not modelled |
70.5 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: class ii fructose-1,6-bisphosphate aldolase from helicobacter pylori2 in complex with phosphoglycolohydroxamic acid, a competitive3 inhibitor
|
|
|
| 102 | d1f74a_ |
|
not modelled |
70.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 103 | c3e96B_ |
|
not modelled |
70.0 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bacillus2 clausii
|
|
|
| 104 | c2cw6B_ |
|
not modelled |
69.7 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa lyase, mitochondrial;
PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
|
|
|
| 105 | c4ah7C_ |
|
not modelled |
69.6 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: structure of wild type stapylococcus aureus n-acetylneuraminic acid2 lyase in complex with pyruvate
|
|
|
| 106 | d1ad1a_ |
|
not modelled |
69.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
| 107 | c5dlcC_ |
|
not modelled |
68.5 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: x-ray crystal structure of a pyridoxine 5-prime-phosphate synthase2 from pseudomonas aeruginosa
|
|
|
| 108 | c5kinC_ |
|
not modelled |
67.6 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
|
|
|
| 109 | c3iwpK_ |
|
not modelled |
67.5 |
16 |
PDB header:metal binding protein
Chain: K: PDB Molecule:copper homeostasis protein cutc homolog;
PDBTitle: crystal structure of human copper homeostasis protein cutc
|
|
|
| 110 | d2gjpa2 |
|
not modelled |
67.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 111 | d1bxca_ |
|
not modelled |
67.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 112 | d3dhpa2 |
|
not modelled |
66.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 113 | c2hmcA_ |
|
not modelled |
66.2 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
|
|
| 114 | c4nq1B_ |
|
not modelled |
66.0 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: legionella pneumophila dihydrodipicolinate synthase with first2 substrate pyruvate bound in the active site
|
|
|
| 115 | c3na8A_ |
|
not modelled |
65.9 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
|
|
| 116 | c4ur7B_ |
|
not modelled |
65.7 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:keto-deoxy-d-galactarate dehydratase;
PDBTitle: crystal structure of keto-deoxy-d-galactarate dehydratase2 complexed with pyruvate
|
|
|
| 117 | c3rxzA_ |
|
not modelled |
65.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:polysaccharide deacetylase;
PDBTitle: crystal structure of putative polysaccharide deacetylase from2 mycobacterium smegmatis
|
|
|
| 118 | c4uxdC_ |
|
not modelled |
65.7 |
9 |
PDB header:lyase
Chain: C: PDB Molecule:2-dehydro-3-deoxy-d-gluconate/2-dehydro-3-deoxy-
PDBTitle: 2-keto 3-deoxygluconate aldolase from picrophilus torridus
|
|
|
| 119 | c3gk0H_ |
|
not modelled |
65.6 |
23 |
PDB header:transferase
Chain: H: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein from2 burkholderia pseudomallei
|
|
|
| 120 | c3b8iF_ |
|
not modelled |
65.5 |
13 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
|
|