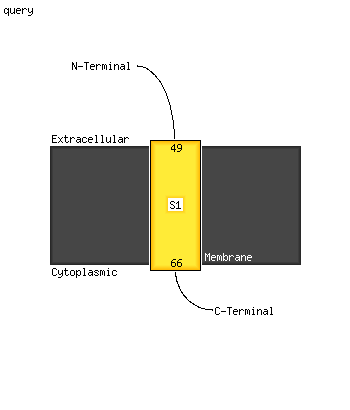
1 c5ex1E_
100.0
21
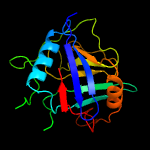
PDB header: isomeraseChain: E: PDB Molecule: peptidyl-prolyl cis-trans isomerase cyclophilin type;PDBTitle: crystal structure of cyclophilin aquacyp300 from hirschia baltica
2 c4r3fA_
100.0
30
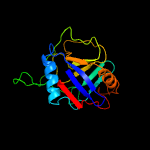
PDB header: isomeraseChain: A: PDB Molecule: spliceosomal protein cwc27;PDBTitle: structure of the spliceosomal peptidyl-prolyl cis-trans isomerase2 cwc27 from chaetomium thermophilum
3 c5ex2A_
100.0
22
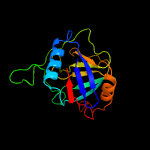
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of cyclophilin aquacyp293 from hirschia baltica
4 c2k7nA_
100.0
31
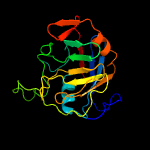
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase-like 1;PDBTitle: solution structure of the ppil1 bound to a fragment of skip
5 c3rfyA_
100.0
28
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase cyp38, chloroplastic;PDBTitle: crystal structure of arabidopsis thaliana cyclophilin 38 (atcyp38)
6 d2a2na1
100.0
31
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
7 c3bo7D_
100.0
24
PDB header: isomerase/immunosuppressantChain: D: PDB Molecule: peptidyl-prolyl cis-trans isomerase cyclophilin-type;PDBTitle: crystal structure of toxoplasma gondii peptidyl-prolyl cis-trans2 isomerase, 541.m00136
8 d2fu0a1
100.0
30
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
9 d1zkca1
100.0
33
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
10 c1ihgA_
100.0
27
PDB header: isomeraseChain: A: PDB Molecule: cyclophilin 40;PDBTitle: bovine cyclophilin 40, monoclinic form
11 c2hq6A_
100.0
29
PDB header: isomeraseChain: A: PDB Molecule: serologically defined colon cancer antigen 10;PDBTitle: structure of the cyclophilin_cecyp16-like domain of the serologically2 defined colon cancer antigen 10 from homo sapiens
12 d2b71a1
100.0
32
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
13 c2b71A_
100.0
32
PDB header: isomeraseChain: A: PDB Molecule: cyclophilin-like protein;PDBTitle: plasmodium yoelii cyclophilin-like protein
14 d1xwna1
100.0
31
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
15 c5jheA_
100.0
28
PDB header: chaperoneChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase cyp7;PDBTitle: the crystal structure of the saccharomyces cerevisiae co-chaperone2 cpr7
16 d1w74a_
100.0
39
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
17 d1ihga2
100.0
27
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
18 d2ok3a1
100.0
35
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
19 d1v9ta_
100.0
30
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
20 c3s6mA_
100.0
28
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: the structure of a peptidyl-prolyl cis-trans isomerase from2 burkholderia pseudomallei
21 c2qerA_
not modelled
100.0
28
PDB header: isomeraseChain: A: PDB Molecule: cyclophilin-like protein, putative;PDBTitle: crystal structure of cryptosporidium parvum cyclophilin type peptidyl-2 prolyl cis-trans isomerase cgd2_1660 in the presence of dipeptide3 ala-pro
22 d1xo7a_
not modelled
100.0
29
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
23 c3jb9d_
not modelled
100.0
32
PDB header: rna binding protein/rnaChain: D: PDB Molecule: small nuclear ribonucleoprotein sm d3;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
24 d1lopa_
not modelled
100.0
29
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
25 d1z81a1
not modelled
100.0
29
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
26 c1z81A_
not modelled
100.0
29
PDB header: isomeraseChain: A: PDB Molecule: cyclophilin;PDBTitle: crystal structure of cyclophilin from plasmodium yoelii.
27 c2poyB_
not modelled
100.0
29
PDB header: isomerase/immunosuppressantChain: B: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans2 isomerase cgd2_4120 in complex with cyclosporin a
28 c3bkpA_
not modelled
100.0
30
PDB header: isomeraseChain: A: PDB Molecule: cyclophilin;PDBTitle: crystal structure of the toxoplasma gondii cyclophilin, 49.m03261
29 d2rmca_
not modelled
100.0
30
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
30 d2r99a1
not modelled
100.0
33
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
31 c2he9B_
not modelled
100.0
25
PDB header: isomeraseChain: B: PDB Molecule: nk-tumor recognition protein;PDBTitle: structure of the peptidylprolyl isomerase domain of the human nk-2 tumour recognition protein
32 d1cyna_
not modelled
100.0
27
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
33 d2esla1
not modelled
100.0
30
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
34 c3k2cA_
not modelled
100.0
26
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of peptidyl-prolyl cis-trans isomerase from2 encephalitozoon cuniculi at 1.9 a resolution
35 d2cfea1
not modelled
100.0
30
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
36 c2ck1A_
not modelled
100.0
29
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase e;PDBTitle: the structure of oxidised cyclophilin a from s. mansoni
37 d2z6wa1
not modelled
100.0
30
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
38 d2igva1
not modelled
100.0
29
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
39 d1a33a_
not modelled
100.0
29
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
40 d1vdna1
not modelled
100.0
28
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
41 c4i9yC_
not modelled
100.0
29
PDB header: transport proteinChain: C: PDB Molecule: e3 sumo-protein ligase ranbp2;PDBTitle: structure of the c-terminal domain of nup358
42 d1qnga_
not modelled
100.0
29
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
43 d1h0pa_
not modelled
100.0
28
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
44 c5ybaC_
not modelled
100.0
30
PDB header: isomeraseChain: C: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: dimeric cyclophilin from t.vaginalis in complex with myb1 peptide
45 d1w8ma_
not modelled
100.0
28
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
46 d1qoia_
not modelled
100.0
31
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
47 c2mvzA_
not modelled
100.0
35
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: solution structure for cyclophilin a from geobacillus kaustophilus
48 c2oseA_
not modelled
100.0
23
PDB header: isomeraseChain: A: PDB Molecule: probable peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of the mimivirus cyclophilin
49 c5mqfo_
not modelled
100.0
38
PDB header: splicingChain: O: PDB Molecule: crooked neck-like protein 1;PDBTitle: cryo-em structure of a human spliceosome activated for step 2 of2 splicing (c* complex)
50 d2c3ba1
not modelled
100.0
24
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Cyclophilin (peptidylprolyl isomerase)
51 c2nnzA_
not modelled
95.2
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: solution structure of the hypothetical protein af2241 from2 archaeoglobus fulgidus
52 c3kopB_
not modelled
89.2
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein with a cyclophilin-like fold2 (yp_831253.1) from arthrobacter sp. fb24 at 1.90 a resolution
53 d1x7fa1
not modelled
80.7
18
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Outer surface protein, C-terminal domain
54 c2p0oA_
not modelled
74.8
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein duf871;PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
55 c1x7fA_
not modelled
71.6
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: outer surface protein;PDBTitle: crystal structure of an uncharacterized b. cereus protein
56 c5dudA_
not modelled
54.6
16
PDB header: unknown functionChain: A: PDB Molecule: ybgk;PDBTitle: crystal structure of e. coli ybgjk
57 d1zx8a1
not modelled
48.3
18
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: TM1367-like
58 c2khfA_
not modelled
29.5
36
PDB header: antimicrobial proteinChain: A: PDB Molecule: plnj;PDBTitle: plantaricin j in dpc-micelles
59 c3mmlE_
not modelled
29.3
19
PDB header: hydrolaseChain: E: PDB Molecule: allophanate hydrolase subunit 2;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
60 c2khgA_
not modelled
26.4
36
PDB header: antimicrobial proteinChain: A: PDB Molecule: plnj;PDBTitle: plantaricin j in tfe
61 c6oegY_
not modelled
22.8
15
PDB header: translocaseChain: Y: PDB Molecule: type iv secretion system apparatus protein cagx;PDBTitle: structure of cagx from a cryo-em reconstruction of a t4ss
62 c5wstA_
not modelled
22.7
16
PDB header: motor proteinChain: A: PDB Molecule: unconventional myosin-viia;PDBTitle: crystal structure of myo7a sah
63 c2kimA_
not modelled
21.0
17
PDB header: transferaseChain: A: PDB Molecule: o6-methylguanine-dna methyltransferase;PDBTitle: 1.7-mm microcryoprobe solution nmr structure of an o6-methylguanine2 dna methyltransferase family protein from vibrio parahaemolyticus.3 northeast structural genomics consortium target vpr247.
64 c3gx4X_
not modelled
19.7
14
PDB header: dna binding protein/dnaChain: X: PDB Molecule: alkyltransferase-like protein 1;PDBTitle: crystal structure analysis of s. pombe atl in complex with dna
65 c2x5rA_
not modelled
18.0
40
PDB header: viral proteinChain: A: PDB Molecule: hypothetical protein orf126;PDBTitle: crystal structure of the hypothetical protein orf126 from pyrobaculum2 spherical virus
66 d1sfea1
not modelled
17.5
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain
67 c5eurC_
not modelled
16.5
29
PDB header: unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: hypothetical protein sf216 from shigella flexneri 5a m90t
68 c3x27C_
not modelled
15.2
10
PDB header: lyaseChain: C: PDB Molecule: cucumopine synthase;PDBTitle: structure of mcbb in complex with tryptophan
69 d1qnta1
not modelled
13.2
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain
70 c3f5hB_
not modelled
12.1
18
PDB header: protein bindingChain: B: PDB Molecule: type i polyketide synthase pikaiii, type i polyketidePDBTitle: crystal structure of fused docking domains from pikaiii and pikaiv of2 the pikromycin polyketide synthase
71 c2d46A_
not modelled
11.8
18
PDB header: metal transportChain: A: PDB Molecule: calcium channel, voltage-dependent, beta 4PDBTitle: solution structure of the human beta4a-a domain
72 d1x0ha1
not modelled
11.5
21
Fold: RGC domain-likeSuperfamily: RGC domain-likeFamily: RGC domain
73 c5nj8D_
not modelled
11.2
16
PDB header: transcriptionChain: D: PDB Molecule: aryl hydrocarbon receptor nuclear translocator;PDBTitle: structural basis for aryl hydrocarbon receptor mediated gene2 activation
74 c1oqyA_
not modelled
10.1
14
PDB header: replicationChain: A: PDB Molecule: uv excision repair protein rad23 homolog a;PDBTitle: structure of the dna repair protein hhr23a
75 c3oepA_
not modelled
9.9
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha0988;PDBTitle: crystal structure of ttha0988 in space group p43212
76 d1xrsb2
not modelled
9.5
57
Fold: Dodecin subunit-likeSuperfamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domainFamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain
77 c6eu1Q_
not modelled
8.7
29
PDB header: transcriptionChain: Q: PDB Molecule: dna-directed rna polymerase iii subunit rpc7;PDBTitle: rna polymerase iii - open dna complex (oc-pol3)
78 c5zm8A_
not modelled
8.5
19
PDB header: lipid transportChain: A: PDB Molecule: oxysterol-binding protein-related protein 2;PDBTitle: crystal structure of orp2-ord in complex with pi(4,5)p2
79 c1z8yI_
not modelled
7.3
36
PDB header: virusChain: I: PDB Molecule: spike glycoprotein e1;PDBTitle: mapping the e2 glycoprotein of alphaviruses
80 c1z8yO_
not modelled
7.3
36
PDB header: virusChain: O: PDB Molecule: spike glycoprotein e1;PDBTitle: mapping the e2 glycoprotein of alphaviruses
81 c1z8yK_
not modelled
7.3
36
PDB header: virusChain: K: PDB Molecule: spike glycoprotein e1;PDBTitle: mapping the e2 glycoprotein of alphaviruses
82 c1z8yM_
not modelled
7.3
36
PDB header: virusChain: M: PDB Molecule: spike glycoprotein e1;PDBTitle: mapping the e2 glycoprotein of alphaviruses
83 c2kncB_
not modelled
7.3
17
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
84 c5z08D_
not modelled
6.5
29
PDB header: cell cycleChain: D: PDB Molecule: cenp-h;PDBTitle: the crystal structure of kinetochore subunits cenp-h/i/k triple2 complex
85 d1m4ma_
not modelled
6.5
24
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat
86 c2j98A_
not modelled
6.3
21
PDB header: rna binding proteinChain: A: PDB Molecule: replicase polyprotein 1ab;PDBTitle: human coronavirus 229e non structural protein 9 cys69ala mutant (nsp9)
87 c2m1aA_
not modelled
6.3
42
PDB header: viral proteinChain: A: PDB Molecule: hiv-1 rev arginine-rich motif (arm);PDBTitle: hiv-1 rev arm peptide (residues t34-r50)
88 c2vdaA_
not modelled
6.3
29
PDB header: protein transportChain: A: PDB Molecule: translocase subunit seca;PDBTitle: solution structure of the seca-signal peptide complex
89 c4uaqA_
not modelled
6.0
39
PDB header: protein transportChain: A: PDB Molecule: protein translocase subunit seca 2;PDBTitle: crystal structure of the accessory translocation atpase, seca2, from2 mycobacterium tuberculosis
90 c6qbzA_
not modelled
5.9
26
PDB header: ribosomal proteinChain: A: PDB Molecule: ribosome hibernation promoting factor;PDBTitle: solution structure of the n-terminal domain of the staphylococcus2 aureus hibernation promoting factor
91 c1kuzB_
not modelled
5.9
33
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: solution structure of the membrane proximal regions of2 alpha-iib and beta-3 integrins
92 c5mmjy_
not modelled
5.8
9
PDB header: ribosomeChain: Y: PDB Molecule: PDBTitle: structure of the small subunit of the chloroplast ribosome
93 c2rqlA_
not modelled
5.7
30
PDB header: translationChain: A: PDB Molecule: probable sigma-54 modulation protein;PDBTitle: solution structure of the e. coli ribosome hibernation2 promoting factor hpf
94 c3m0aD_
not modelled
5.6
55
PDB header: signaling proteinChain: D: PDB Molecule: baculoviral iap repeat-containing protein 3;PDBTitle: crystal structure of traf2:ciap2 complex
95 c4rgwA_
not modelled
5.6
37
PDB header: transferase/transcriptionChain: A: PDB Molecule: transcription initiation factor tfiid subunit 1;PDBTitle: crystal structure of a taf1-taf7 complex in human transcription factor2 iid
96 d2qfaa1
not modelled
5.5
29
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat
97 d1l4sa_
not modelled
5.3
35
Fold: Ribosome binding protein Y (YfiA homologue)Superfamily: Ribosome binding protein Y (YfiA homologue)Family: Ribosome binding protein Y (YfiA homologue)
98 d1imua_
not modelled
5.2
33
Fold: Ribosome binding protein Y (YfiA homologue)Superfamily: Ribosome binding protein Y (YfiA homologue)Family: Ribosome binding protein Y (YfiA homologue)
99 c4zyeA_
not modelled
5.0
27
PDB header: transferaseChain: A: PDB Molecule: methylated-dna--protein-cysteine methyltransferase;PDBTitle: crystal structure of sulfolobus solfataricus o6-methylguanine2 methyltransferase