| 1 |
|
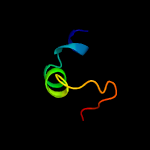
PDB 4mkq chain A
Region: 102 - 116
Aligned: 15
Modelled: 15
Confidence: 35.4%
Identity: 7%
PDB header:toxin
Chain: A: PDB Molecule:monalysin;
PDBTitle: crystal structure of the pore-forming toxin monalysin mutant deleted2 of the membrane-spanning domain
Phyre2
| 2 |
|
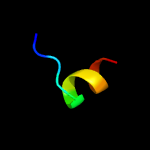
PDB 4mno chain A
Region: 27 - 39
Aligned: 13
Modelled: 13
Confidence: 25.4%
Identity: 54%
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor 1a;
PDBTitle: crystal structure of aif1a from pyrococcus abyssi
Phyre2
| 3 |
|
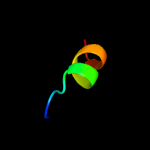
PDB 1yce chain D
Region: 38 - 63
Aligned: 26
Modelled: 26
Confidence: 20.9%
Identity: 31%
PDB header:membrane protein
Chain: D: PDB Molecule:subunit c;
PDBTitle: structure of the rotor ring of f-type na+-atpase from ilyobacter2 tartaricus
Phyre2
| 4 |
|
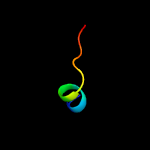
PDB 2oqk chain A
Region: 26 - 39
Aligned: 14
Modelled: 14
Confidence: 18.6%
Identity: 21%
PDB header:translation
Chain: A: PDB Molecule:putative translation initiation factor eif-1a;
PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
Phyre2
| 5 |
|
PDB 1vw4 chain 8
Region: 20 - 45
Aligned: 24
Modelled: 26
Confidence: 17.8%
Identity: 17%
PDB header:ribosome
Chain: 8: PDB Molecule:54s ribosomal protein l13, mitochondrial;
PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
Phyre2
| 6 |
|
PDB 5e06 chain A
Region: 23 - 34
Aligned: 12
Modelled: 12
Confidence: 16.9%
Identity: 33%
PDB header:nuclear protein
Chain: A: PDB Molecule:nucleocapsid protein;
PDBTitle: structure of sin nombre virus nucleoprotein in long-axis crystal form
Phyre2
| 7 |
|
PDB 2dgy chain A
Region: 27 - 39
Aligned: 13
Modelled: 13
Confidence: 16.0%
Identity: 31%
PDB header:translation
Chain: A: PDB Molecule:mgc11102 protein;
PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
Phyre2
| 8 |
|
PDB 1jt8 chain A
Region: 26 - 38
Aligned: 13
Modelled: 13
Confidence: 14.2%
Identity: 23%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 9 |
|
PDB 5dn6 chain P
Region: 38 - 63
Aligned: 26
Modelled: 26
Confidence: 13.7%
Identity: 27%
PDB header:hydrolase
Chain: P: PDB Molecule:atp synthase f0 subcomplex c subunit;
PDBTitle: atp synthase from paracoccus denitrificans
Phyre2
| 10 |
|
PDB 4mjt chain H
Region: 102 - 116
Aligned: 15
Modelled: 15
Confidence: 13.1%
Identity: 7%
PDB header:toxin
Chain: H: PDB Molecule:monalysin;
PDBTitle: crystal structure of the oligomeric pore-forming toxin pro-monalysin
Phyre2
| 11 |
|
PDB 4v1g chain A
Region: 38 - 63
Aligned: 26
Modelled: 26
Confidence: 12.1%
Identity: 23%
PDB header:hydrolase
Chain: A: PDB Molecule:f0f1 atp synthase subunit c;
PDBTitle: crystal structure of a mycobacterial atp synthase rotor ring
Phyre2
| 12 |
|
PDB 5fsg chain A
Region: 23 - 34
Aligned: 12
Modelled: 12
Confidence: 11.1%
Identity: 42%
PDB header:viral protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein, hantavirus
PDBTitle: structure of the hantavirus nucleoprotein provides insights2 into the mechanism of rna encapsidation and a template for3 drug design
Phyre2
| 13 |
|
PDB 4bpp chain 0
Region: 27 - 38
Aligned: 12
Modelled: 12
Confidence: 10.1%
Identity: 33%
PDB header:ribosome
Chain: 0: PDB Molecule:translation initiation factor eif-1a family protein;
PDBTitle: the crystal structure of the eukaryotic 40s ribosomal subunit in2 complex with eif1 and eif1a - complex 4
Phyre2
| 14 |
|
PDB 6f36 chain E
Region: 35 - 63
Aligned: 29
Modelled: 29
Confidence: 9.3%
Identity: 21%
PDB header:proton transport
Chain: E: PDB Molecule:mitochondrial atp synthase subunit c;
PDBTitle: polytomella fo model
Phyre2
| 15 |
|
PDB 4rnd chain B
Region: 8 - 52
Aligned: 45
Modelled: 45
Confidence: 9.2%
Identity: 18%
PDB header:hydrolase
Chain: B: PDB Molecule:v-type proton atpase subunit f;
PDBTitle: crystal structure of the subunit df-assembly of the eukaryotic v-2 atpase.
Phyre2
| 16 |
|
PDB 2w5j chain M
Region: 38 - 63
Aligned: 26
Modelled: 26
Confidence: 8.6%
Identity: 31%
PDB header:hydrolase
Chain: M: PDB Molecule:atp synthase c chain, chloroplastic;
PDBTitle: structure of the c14-rotor ring of the proton translocating2 chloroplast atp synthase
Phyre2
| 17 |
|
PDB 3fbz chain A
Region: 29 - 39
Aligned: 11
Modelled: 11
Confidence: 7.5%
Identity: 18%
PDB header:structural protein
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of orf140 of the archaeal virus acidianus2 filamentous virus 1 (afv1)
Phyre2
| 18 |
|
PDB 1d7q chain A
Region: 26 - 39
Aligned: 14
Modelled: 14
Confidence: 6.7%
Identity: 36%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 19 |
|
PDB 2vy2 chain A
Region: 24 - 48
Aligned: 24
Modelled: 25
Confidence: 5.7%
Identity: 46%
PDB header:transcription
Chain: A: PDB Molecule:protein leafy;
PDBTitle: structure of leafy transcription factor from arabidopsis2 thaliana in complex with dna from ag-i promoter
Phyre2
| 20 |
|
PDB 2lde chain A
Region: 34 - 40
Aligned: 7
Modelled: 6
Confidence: 5.3%
Identity: 14%
PDB header:toxin
Chain: A: PDB Molecule:sarafotoxin-i3;
PDBTitle: solution structure of the long sarafotoxin srtx-i3
Phyre2
| 21 |
|