1 c3anoA_
100.0
100
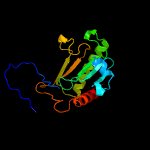
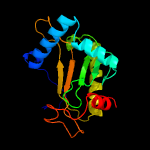
PDB header: transferaseChain: A: PDB Molecule: ap-4-a phosphorylase;PDBTitle: crystal structure of a novel diadenosine 5',5'''-p1,p4-tetraphosphate2 phosphorylase from mycobacterium tuberculosis h37rv
2 c3ksvA_
100.0
21
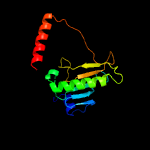
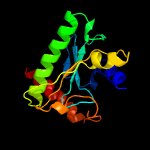
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: hypothetical protein from leishmania major
3 c3l7xA_
100.0
23
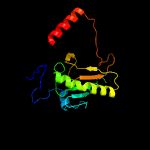
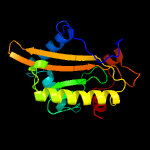
PDB header: cell cycleChain: A: PDB Molecule: putative hit-like protein involved in cell-cyclePDBTitle: the crystal structure of smu.412c from streptococcus mutans ua159
4 c3imiB_
100.0
23
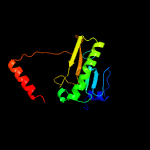
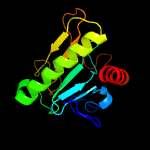
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hit family protein;PDBTitle: 2.01 angstrom resolution crystal structure of a hit family protein2 from bacillus anthracis str. 'ames ancestor'
5 d1y23a_
100.0
25
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
6 c2eo4A_
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: 150aa long hypothetical histidine triad nucleotide-bindingPDBTitle: crystal structure of hypothetical histidine triad nucleotide-binding2 protein st2152 from sulfolobus tokodaii strain7
7 c6iq1A_
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: adenosine 5'-monophosphoramidase;PDBTitle: crystal structure of histidine triad nucleotide-binding protein from2 candida albicans
8 c3lb5B_
100.0
21
PDB header: cell cycleChain: B: PDB Molecule: hit-like protein involved in cell-cycle regulation;PDBTitle: crystal structure of hit-like protein involved in cell-cycle2 regulation from bartonella henselae with unknown ligand
9 c5cs2A_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: histidine triad protein;PDBTitle: crystal structure of plasmodium falciparum diadenosine triphosphate2 hydrolase in complex with cyclomarin a
10 c1emsB_
100.0
24
PDB header: antitumor proteinChain: B: PDB Molecule: nit-fragile histidine triad fusion protein;PDBTitle: crystal structure of the c. elegans nitfhit protein
11 c3o0mB_
100.0
24
PDB header: hydrolaseChain: B: PDB Molecule: hit family protein;PDBTitle: crystal structure of a zn-bound histidine triad family protein from2 mycobacterium smegmatis
12 c1zwjA_
100.0
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative galactose-1-phosphate uridyl transferase;PDBTitle: x-ray structure of galt-like protein from arabidopsis thaliana2 at5g18200
13 c1gupC_
100.0
16
PDB header: nucleotidyltransferaseChain: C: PDB Molecule: galactose-1-phosphate uridylyltransferase;PDBTitle: structure of nucleotidyltransferase complexed with udp-2 galactose
14 c3p0tB_
100.0
25
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an hit-like protein from mycobacterium2 paratuberculosis
15 c5in3A_
100.0
15
PDB header: transferaseChain: A: PDB Molecule: galactose-1-phosphate uridylyltransferase;PDBTitle: crystal structure of glucose-1-phosphate bound nucleotidylated human2 galactose-1-phosphate uridylyltransferase
16 d1emsa1
100.0
24
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
17 d1z84a2
100.0
14
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase
18 d1guqa2
100.0
16
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase
19 d1fita_
100.0
30
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
20 d2oika1
100.0
25
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
21 c3r6fA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: hit family protein;PDBTitle: crystal structure of a zinc-containing hit family protein from2 encephalitozoon cuniculi
22 d1rzya_
not modelled
100.0
18
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
23 d1kpfa_
not modelled
100.0
19
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
24 c3n1tE_
not modelled
100.0
18
PDB header: hydrolaseChain: E: PDB Molecule: hit-like protein hint;PDBTitle: crystal structure of the h101a mutant echint gmp complex
25 c4eguA_
not modelled
99.9
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: histidine triad (hit) protein;PDBTitle: 0.95a resolution structure of a histidine triad protein from2 clostridium difficile
26 d1xqua_
not modelled
99.9
19
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
27 c1xquA_
not modelled
99.9
19
PDB header: hydrolaseChain: A: PDB Molecule: hit family hydrolase;PDBTitle: hit family hydrolase from clostridium thermocellum cth-393
28 c6d6jB_
not modelled
99.9
22
PDB header: hydrolaseChain: B: PDB Molecule: hit family hydrolase;PDBTitle: crystal structure of hit family hydrolase from legionella pneumophila2 philadelphia 1
29 c3oj7A_
not modelled
99.9
25
PDB header: metal binding proteinChain: A: PDB Molecule: putative histidine triad family protein;PDBTitle: crystal structure of a histidine triad family protein from entamoeba2 histolytica, bound to sulfate
30 c3i24B_
not modelled
99.9
18
PDB header: hydrolaseChain: B: PDB Molecule: hit family hydrolase;PDBTitle: crystal structure of a hit family hydrolase protein from vibrio2 fischeri. northeast structural genomics consortium target id vfr176
31 c4q61J_
not modelled
99.9
19
PDB header: cell cycleChain: J: PDB Molecule: uncharacterized hit-like protein hp_0404;PDBTitle: hit like protein from helicobacter pylori 26695
32 c3oheA_
not modelled
99.9
16
PDB header: hydrolaseChain: A: PDB Molecule: histidine triad (hit) protein;PDBTitle: crystal structure of a histidine triad protein (maqu_1709) from2 marinobacter aquaeolei vt8 at 1.20 a resolution
33 c4incA_
not modelled
99.9
18
PDB header: hydrolaseChain: A: PDB Molecule: histidine triad nucleotide-binding protein 2,PDBTitle: human histidine triad nucleotide binding protein 2
34 c3nrdB_
not modelled
99.9
27
PDB header: nucleotide binding proteinChain: B: PDB Molecule: histidine triad (hit) protein;PDBTitle: crystal structure of a histidine triad (hit) protein (smc02904) from2 sinorhizobium meliloti 1021 at 2.06 a resolution
35 c3i4sB_
not modelled
99.9
19
PDB header: hydrolaseChain: B: PDB Molecule: histidine triad protein;PDBTitle: crystal structure of histidine triad protein blr8122 from2 bradyrhizobium japonicum
36 d1z84a1
not modelled
99.8
22
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase
37 c4ndgB_
not modelled
99.7
22
PDB header: dna binding protein/rna/dnaChain: B: PDB Molecule: aprataxin;PDBTitle: human aprataxin (aptx) bound to rna-dna and zn - adenosine vanadate2 transition state mimic complex
38 c3splC_
not modelled
99.5
15
PDB header: hydrolase/dnaChain: C: PDB Molecule: aprataxin-like protein;PDBTitle: crystal structure of aprataxin ortholog hnt3 in complex with dna and2 amp
39 c3jb9c_
not modelled
99.5
14
PDB header: rna binding protein/rnaChain: C: PDB Molecule: u5 snrna;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
40 c4i5wA_
not modelled
99.5
23
PDB header: transferaseChain: A: PDB Molecule: 5',5'''-p-1,p-4-tetraphosphate phosphorylase 2;PDBTitle: crystal structure of yeast ap4a phosphorylase apa2 in complex with amp
41 c4qvuA_
not modelled
99.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf4931 family protein (bce0241) from bacillus2 cereus atcc 10987 at 2.65 a resolution
42 c6id1U_
not modelled
99.1
18
PDB header: splicingChain: U: PDB Molecule: cwf19-like protein 2;PDBTitle: cryo-em structure of a human intron lariat spliceosome after prp432 loaded (ils2 complex) at 2.9 angstrom resolution
43 d3bl9a1
not modelled
98.9
25
Fold: HIT-likeSuperfamily: HIT-likeFamily: mRNA decapping enzyme DcpS C-terminal domain
44 c3bl9B_
not modelled
98.7
25
PDB header: hydrolaseChain: B: PDB Molecule: scavenger mrna-decapping enzyme dcps;PDBTitle: synthetic gene encoded dcps bound to inhibitor dg157493
45 d1vlra1
not modelled
98.6
23
Fold: HIT-likeSuperfamily: HIT-likeFamily: mRNA decapping enzyme DcpS C-terminal domain
46 d1guqa1
not modelled
98.5
13
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase
47 c1xmlA_
not modelled
98.5
22
PDB header: chaperoneChain: A: PDB Molecule: heat shock-like protein 1;PDBTitle: structure of human dcps
48 c5bv3C_
not modelled
98.4
17
PDB header: hydrolaseChain: C: PDB Molecule: m7gpppx diphosphatase;PDBTitle: yeast scavenger decapping enzyme in complex with m7gdp
49 c6gbsB_
not modelled
98.3
24
PDB header: hydrolaseChain: B: PDB Molecule: putative mrna decapping protein;PDBTitle: crystal structure of the c. themophilum scavenger decapping enzyme2 dcps apo form
50 c4lvjA_
not modelled
92.6
17
PDB header: dna binding protein/dnaChain: A: PDB Molecule: plasmid recombination enzyme;PDBTitle: mobm relaxase domain (mobv; mob_pre) bound to plasmid pmv158 orit dna2 (22nt). mn-bound crystal structure at ph 5.5
51 d2pofa1
not modelled
90.2
12
Fold: HIT-likeSuperfamily: HIT-likeFamily: CDH-like
52 d1mg4a_
not modelled
49.8
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: Doublecortin (DC)Family: Doublecortin (DC)
53 c5kciA_
not modelled
49.8
26
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein ypl067c;PDBTitle: crystal structure of htc1
54 d1uf0a_
not modelled
47.3
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: Doublecortin (DC)Family: Doublecortin (DC)
55 c4atuI_
not modelled
38.4
22
PDB header: hydrolaseChain: I: PDB Molecule: neuronal migration protein doublecortin;PDBTitle: human doublecortin n-dc repeat plus linker, and tubulin (2xrp) docked2 into an 8a cryo-em map of doublecortin-stabilised microtubules3 reconstructed in absence of kinesin
56 d1mjda_
not modelled
30.1
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: Doublecortin (DC)Family: Doublecortin (DC)
57 c2dnfA_
not modelled
27.1
11
PDB header: protein bindingChain: A: PDB Molecule: doublecortin domain-containing protein 2;PDBTitle: solution structure of rsgi ruh-062, a dcx domain from human
58 c1vr7A_
not modelled
26.6
15
PDB header: lyaseChain: A: PDB Molecule: s-adenosylmethionine decarboxylase proenzyme;PDBTitle: crystal structure of s-adenosylmethionine decarboxylase proenzyme2 (tm0655) from thermotoga maritima at 1.2 a resolution
59 c5ip4E_
not modelled
26.4
12
PDB header: transferaseChain: E: PDB Molecule: neuronal migration protein doublecortin;PDBTitle: x-ray structure of the c-terminal domain of human doublecortin
60 d1vr7a1
not modelled
25.5
15
Fold: S-adenosylmethionine decarboxylaseSuperfamily: S-adenosylmethionine decarboxylaseFamily: Bacterial S-adenosylmethionine decarboxylase
61 c2v4xA_
not modelled
23.8
15
PDB header: viral proteinChain: A: PDB Molecule: capsid protein p27;PDBTitle: crystal structure of jaagsiekte sheep retrovirus capsid n-2 terminal domain
62 c6f21B_
not modelled
23.8
24
PDB header: toxinChain: B: PDB Molecule: dendroaspis polylepis mt9;PDBTitle: crystal structure of toxin mt9 from mamba venom
63 c1ayeA_
not modelled
22.3
19
PDB header: serine proteaseChain: A: PDB Molecule: procarboxypeptidase a2;PDBTitle: human procarboxypeptidase a2
64 d1tfsa_
not modelled
21.4
25
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins
65 d1ntxa_
not modelled
20.9
25
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins
66 d1v76a_
not modelled
20.7
17
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: RNase P subunit p29-like
67 c5mm2A_
not modelled
19.4
30
PDB header: virusChain: A: PDB Molecule: capsid protein vp4c;PDBTitle: nora virus structure
68 c2zaeC_
not modelled
18.8
17
PDB header: hydrolaseChain: C: PDB Molecule: ribonuclease p protein component 1;PDBTitle: crystal structure of protein ph1601p in complex with protein ph1771p2 of archaeal ribonuclease p from pyrococcus horikoshii ot3
69 c5lxjA_
not modelled
18.3
29
PDB header: viral proteinChain: A: PDB Molecule: phosphoprotein;PDBTitle: solution nmr structure of the x domain of peste des petits ruminants2 phosphoprotein
70 d1oqka_
not modelled
17.6
16
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: RNase P subunit p29-like
71 c5j1mD_
not modelled
17.3
40
PDB header: hydrolaseChain: D: PDB Molecule: toxr-activated gene (tage);PDBTitle: crystal structure of csd1-csd2 dimer ii
72 c2qwwB_
not modelled
17.1
32
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of multiple antibiotic-resistance repressor (marr)2 (yp_013417.1) from listeria monocytogenes 4b f2365 at 2.07 a3 resolution
73 d1iq9a_
not modelled
17.0
29
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins
74 c6k0aF_
not modelled
16.6
22
PDB header: rna binding protein/rnaChain: F: PDB Molecule: ribonuclease p protein component 1;PDBTitle: cryo-em structure of an archaeal ribonuclease p
75 d1tsfa_
not modelled
16.3
20
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: RNase P subunit p29-like
76 d1b77a2
not modelled
16.2
13
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor
77 d3ebxa_
not modelled
15.9
29
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins
78 c6ahrD_
not modelled
14.7
14
PDB header: hydrolase/rnaChain: D: PDB Molecule: ribonuclease p protein subunit p29;PDBTitle: cryo-em structure of human ribonuclease p
79 c3csqC_
not modelled
14.4
20
PDB header: hydrolaseChain: C: PDB Molecule: morphogenesis protein 1;PDBTitle: crystal and cryoem structural studies of a cell wall2 degrading enzyme in the bacteriophage phi29 tail
80 c3txxD_
not modelled
13.4
28
PDB header: transferaseChain: D: PDB Molecule: putrescine carbamoyltransferase;PDBTitle: crystal structure of putrescine transcarbamylase from enterococcus2 faecalis
81 d1qm7a_
not modelled
13.3
23
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins
82 c3bdeA_
not modelled
13.1
10
PDB header: unknown functionChain: A: PDB Molecule: mll5499 protein;PDBTitle: crystal structure of a dabb family protein with a ferredoxin-like fold2 (mll5499) from mesorhizobium loti maff303099 at 1.79 a resolution
83 c5b0hB_
not modelled
12.6
24
PDB header: metal binding proteinChain: B: PDB Molecule: leukocyte cell-derived chemotaxin-2;PDBTitle: crystal structure of human leukocyte cell-derived chemotaxin 2
84 d1vb0a_
not modelled
12.5
42
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins
85 d1vlva1
not modelled
12.2
17
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase
86 d1jb3a_
not modelled
12.2
30
Fold: OB-foldSuperfamily: TIMP-likeFamily: The laminin-binding domain of agrin
87 c2aydA_
not modelled
11.8
21
PDB header: transcriptionChain: A: PDB Molecule: wrky transcription factor 1;PDBTitle: crystal structure of the c-terminal wrky domainof atwrky1, an sa-2 induced and partially npr1-dependent transcription factor
88 c4agrB_
not modelled
11.7
26
PDB header: sugar binding proteinChain: B: PDB Molecule: galectin;PDBTitle: structure of a tetrameric galectin from cinachyrella sp. (ball2 sponge)
89 c6b4aB_
not modelled
11.4
18
PDB header: structural proteinChain: B: PDB Molecule: doublecortin;PDBTitle: crystal structure of the c-terminal domain of doublecortin (tgdcx)2 from toxoplasma gondii me49
90 d1g6ma_
not modelled
11.2
23
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins
91 d1pc0a_
not modelled
10.7
20
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: RNase P subunit p29-like
92 d1v6pa_
not modelled
10.6
23
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins
93 c3fmbA_
not modelled
10.5
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: dimeric protein of unknown function and ferredoxin-likePDBTitle: crystal structure of dimeric protein of unknown function and2 ferredoxin-like fold (yp_212648.1) from bacteroides fragilis nctc3 9343 at 1.85 a resolution
94 d1dxha1
not modelled
10.5
17
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase
95 d1wiia_
not modelled
10.4
17
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Putative zinc binding domain
96 c6q8iK_
not modelled
10.2
50
PDB header: splicingChain: K: PDB Molecule: protein red;PDBTitle: nterminal domain of human smu1 in complex with human redmid
97 c2yv9B_
not modelled
10.2
12
PDB header: metal transportChain: B: PDB Molecule: chloride intracellular channel exc-4;PDBTitle: crystal structure of the clic homologue exc-4 from c.2 elegans
98 d1ml4a1
not modelled
10.1
20
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase
99 c5op0B_
not modelled
9.9
21
PDB header: transferaseChain: B: PDB Molecule: dna polymerase ligd, polymerase domain;PDBTitle: structure of prim-polc from mycobacterium smegmatis