| 1 |
|
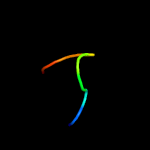
PDB 5e9o chain C
Region: 47 - 68
Aligned: 22
Modelled: 22
Confidence: 20.1%
Identity: 36%
PDB header:hydrolase
Chain: C: PDB Molecule:cellulase, glycosyl hydrolase family 5, tps linker, domain
PDBTitle: spirochaeta thermophila x module - cbm64 - mutant g504a
Phyre2
| 2 |
|
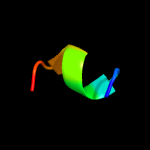
PDB 2fd5 chain A domain 2
Region: 116 - 137
Aligned: 22
Modelled: 22
Confidence: 16.9%
Identity: 45%
Fold: Tetracyclin repressor-like, C-terminal domain
Superfamily: Tetracyclin repressor-like, C-terminal domain
Family: Tetracyclin repressor-like, C-terminal domain
Phyre2
| 3 |
|
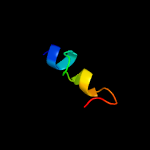
PDB 2mpn chain B
Region: 72 - 101
Aligned: 30
Modelled: 30
Confidence: 12.4%
Identity: 23%
PDB header:membrane protein
Chain: B: PDB Molecule:inner membrane protein ygap;
PDBTitle: 3d nmr structure of the transmembrane domain of the full-length inner2 membrane protein ygap from escherichia coli
Phyre2
| 4 |
|
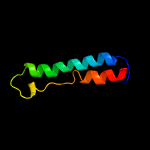
PDB 1cfg chain A
Region: 48 - 57
Aligned: 10
Modelled: 10
Confidence: 11.4%
Identity: 50%
PDB header:coagulation factor
Chain: A: PDB Molecule:coagulation factor viii;
PDBTitle: membrane-binding peptide from the c2 domain of factor viii2 forms an amphipathic structure as determined by nmr3 spectroscopy
Phyre2
| 5 |
|
PDB 6n0t chain A
Region: 38 - 45
Aligned: 8
Modelled: 8
Confidence: 8.5%
Identity: 75%
PDB header:ligase
Chain: A: PDB Molecule:trna ligase;
PDBTitle: trna ligase
Phyre2
| 6 |
|
PDB 2mpn chain A
Region: 77 - 101
Aligned: 25
Modelled: 25
Confidence: 7.9%
Identity: 24%
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein ygap;
PDBTitle: 3d nmr structure of the transmembrane domain of the full-length inner2 membrane protein ygap from escherichia coli
Phyre2
| 7 |
|
PDB 2n5n chain A
Region: 16 - 75
Aligned: 59
Modelled: 60
Confidence: 7.5%
Identity: 20%
PDB header:dna binding protein
Chain: A: PDB Molecule:chromodomain-helicase-dna-binding protein 4;
PDBTitle: structure of an n-terminal domain of chd4
Phyre2
| 8 |
|
PDB 1q0q chain A domain 3
Region: 46 - 54
Aligned: 8
Modelled: 9
Confidence: 7.3%
Identity: 50%
Fold: FwdE/GAPDH domain-like
Superfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family: Dihydrodipicolinate reductase-like
Phyre2
| 9 |
|
PDB 4uzn chain A
Region: 49 - 55
Aligned: 7
Modelled: 7
Confidence: 6.9%
Identity: 43%
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-1,4-glucanase (celulase b);
PDBTitle: the native structure of the family 46 carbohydrate-binding2 module (cbm46) of endo-beta-1,4-glucanase b (cel5b) from3 bacillus halodurans
Phyre2
| 10 |
|
PDB 5kqo chain A
Region: 46 - 54
Aligned: 8
Modelled: 9
Confidence: 6.6%
Identity: 50%
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from vibrio vulnificus
Phyre2
| 11 |
|
PDB 2o3b chain B domain 1
Region: 39 - 52
Aligned: 14
Modelled: 14
Confidence: 6.6%
Identity: 50%
Fold: Nuclease A inhibitor (NuiA)
Superfamily: Nuclease A inhibitor (NuiA)
Family: Nuclease A inhibitor (NuiA)
Phyre2
| 12 |
|
PDB 3au9 chain A
Region: 46 - 54
Aligned: 8
Modelled: 9
Confidence: 6.4%
Identity: 63%
PDB header:isomerase/isomerase inhibitor
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of the quaternary complex-1 of an isomerase
Phyre2
| 13 |
|
PDB 2egh chain A
Region: 46 - 54
Aligned: 8
Modelled: 9
Confidence: 6.1%
Identity: 50%
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase2 complexed with a magnesium ion, nadph and fosmidomycin
Phyre2
| 14 |
|
PDB 1a5t chain A domain 1
Region: 42 - 61
Aligned: 17
Modelled: 20
Confidence: 5.9%
Identity: 24%
Fold: post-AAA+ oligomerization domain-like
Superfamily: post-AAA+ oligomerization domain-like
Family: DNA polymerase III clamp loader subunits, C-terminal domain
Phyre2
| 15 |
|
PDB 2mpk chain A
Region: 10 - 62
Aligned: 53
Modelled: 53
Confidence: 5.9%
Identity: 25%
PDB header:transferase
Chain: A: PDB Molecule:chitin synthase 1;
PDBTitle: characterization and structure of the mit1 domain of a chitin synthase2 from the oomycete saprolegnia monoica
Phyre2
| 16 |
|
PDB 1ry3 chain A
Region: 131 - 145
Aligned: 15
Modelled: 15
Confidence: 5.7%
Identity: 47%
PDB header:antibiotic
Chain: A: PDB Molecule:bacteriocin carnobacteriocin b2;
PDBTitle: nmr solution structure of the precursor for2 carnobacteriocin b2, an antimicrobial peptide from3 carnobacterium piscicola
Phyre2
| 17 |
|
PDB 6b3y chain A
Region: 8 - 20
Aligned: 13
Modelled: 13
Confidence: 5.7%
Identity: 54%
PDB header:transport protein
Chain: A: PDB Molecule:denn domain-containing protein 3;
PDBTitle: crystal structure of the ph-like domain from dennd3
Phyre2
| 18 |
|
PDB 3zfi chain B
Region: 53 - 85
Aligned: 33
Modelled: 33
Confidence: 5.6%
Identity: 12%
PDB header:transport protein
Chain: B: PDB Molecule:rap1a protein;
PDBTitle: rap1a protein (sma2260) from serratia marcescens
Phyre2
| 19 |
|
PDB 2lis chain A
Region: 33 - 44
Aligned: 12
Modelled: 12
Confidence: 5.6%
Identity: 25%
Fold: Fertilization protein
Superfamily: Fertilization protein
Family: Fertilization protein
Phyre2
| 20 |
|
PDB 2jo1 chain A
Region: 91 - 112
Aligned: 22
Modelled: 22
Confidence: 5.5%
Identity: 27%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 21 |
|