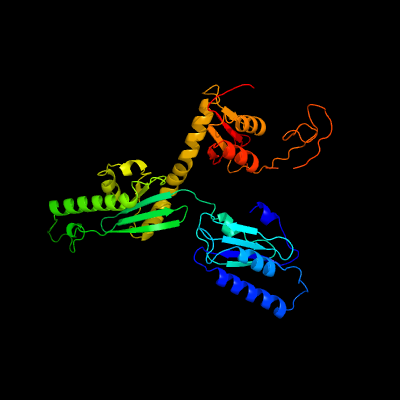
| 1 | c4af1A_
|
|
 |
100.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptide chain release factor subunit 1;
PDBTitle: archeal release factor arf1
|
|
|
|
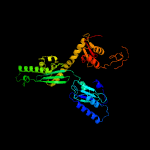
| 2 | c1dt9A_
|
|
 |
100.0 |
17 |
PDB header:translation
Chain: A: PDB Molecule:protein (eukaryotic peptide chain release factor
PDBTitle: the crystal structure of human eukaryotic release factor2 erf1-mechanism of stop codon recognition and peptidyl-trna3 hydrolysis
|
|
|
|
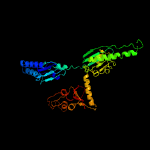
| 3 | c4crnX_
|
|
 |
100.0 |
18 |
PDB header:translation
Chain: X: PDB Molecule:erf1 in ribosome-bound erf1-erf3-gdpnp complex;
PDBTitle: cryo-em of a pretermination complex with erf1 and erf3
|
|
|
|
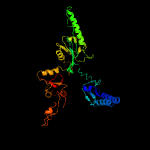
| 4 | c3agkA_
|
|
 |
100.0 |
18 |
PDB header:translation
Chain: A: PDB Molecule:peptide chain release factor subunit 1;
PDBTitle: crystal structure of archaeal translation termination factor, arf1
|
|
|
|
| 5 | c3e1yD_
|
|
 |
100.0 |
19 |
PDB header:translation
Chain: D: PDB Molecule:eukaryotic peptide chain release factor subunit 1;
PDBTitle: crystal structure of human erf1/erf3 complex
|
|
|
|
| 6 | c3mcaB_
|
|
 |
100.0 |
12 |
PDB header:translation regulation/hydrolase
Chain: B: PDB Molecule:protein dom34;
PDBTitle: structure of the dom34-hbs1 complex and implications for its role in2 no-go decay
|
|
|
|
| 7 | c3agjD_
|
|
 |
100.0 |
18 |
PDB header:translation/hydrolase
Chain: D: PDB Molecule:protein pelota homolog;
PDBTitle: crystal structure of archaeal pelota and gtp-bound ef1 alpha complex
|
|
|
|
| 8 | c3agjB_
|
|
 |
100.0 |
18 |
PDB header:translation/hydrolase
Chain: B: PDB Molecule:protein pelota homolog;
PDBTitle: crystal structure of archaeal pelota and gtp-bound ef1 alpha complex
|
|
|
|
| 9 | c3j15A_
|
|
 |
100.0 |
20 |
PDB header:translation/transport protein
Chain: A: PDB Molecule:protein pelota;
PDBTitle: model of ribosome-bound archaeal pelota and abce1
|
|
|
|
| 10 | c3e20C_
|
|
 |
99.9 |
21 |
PDB header:translation
Chain: C: PDB Molecule:eukaryotic peptide chain release factor subunit 1;
PDBTitle: crystal structure of s.pombe erf1/erf3 complex
|
|
|
|
| 11 | c3obwA_
|
|
 |
99.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein pelota homolog;
PDBTitle: crystal structure of two archaeal pelotas reveal inter-domain2 structural plasticity
|
|
|
|
| 12 | c2vgmA_
|
|
 |
99.9 |
14 |
PDB header:cell cycle
Chain: A: PDB Molecule:dom34;
PDBTitle: structure of yeast dom34 : a protein related to translation2 termination factor erf1 and involved in no-go decay.
|
|
|
|
| 13 | c3ir9A_
|
|
 |
99.9 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:peptide chain release factor subunit 1;
PDBTitle: c-terminal domain of peptide chain release factor from methanosarcina2 mazei.
|
|
|
|
| 14 | c3obyB_
|
|
 |
99.9 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein pelota homolog;
PDBTitle: crystal structure of archaeoglobus fulgidus pelota reveals inter-2 domain structural plasticity
|
|
|
|
| 15 | d1dt9a2
|
|
 |
99.8 |
18 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:ERF1/Dom34 C-terminal domain-like |
|
|
|
| 16 | c2qi2A_
|
|
 |
99.8 |
17 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein pelota related protein;
PDBTitle: crystal structure of the thermoplasma acidophilum pelota2 protein
|
|
|
|
| 17 | c2ktvA_
|
|
 |
99.8 |
21 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic peptide chain release factor subunit 1;
PDBTitle: human erf1 c-domain, "open" conformer
|
|
|
|
| 18 | c3e20H_
|
|
 |
99.7 |
26 |
PDB header:translation
Chain: H: PDB Molecule:eukaryotic peptide chain release factor subunit 1;
PDBTitle: crystal structure of s.pombe erf1/erf3 complex
|
|
|
|
| 19 | d1x52a1
|
|
 |
99.5 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:ERF1/Dom34 C-terminal domain-like |
|
|
|
| 20 | d1dt9a1
|
|
 |
99.5 |
19 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:ERF1/Dom34 middle domain-like |
|
|
|
| 21 | d2vgna3 |
|
not modelled |
99.5 |
16 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:ERF1/Dom34 C-terminal domain-like |
|
|
| 22 | d2qi2a3 |
|
not modelled |
99.3 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:ERF1/Dom34 C-terminal domain-like |
|
|
| 23 | d1dt9a3 |
|
not modelled |
99.0 |
11 |
Fold:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Superfamily:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Family:N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 |
|
|
| 24 | d2qi2a2 |
|
not modelled |
97.9 |
17 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:ERF1/Dom34 middle domain-like |
|
|
| 25 | d2vgna2 |
|
not modelled |
96.7 |
10 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:ERF1/Dom34 middle domain-like |
|
|
| 26 | c3on1A_ |
|
not modelled |
76.6 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:bh2414 protein;
PDBTitle: the structure of a protein with unknown function from bacillus2 halodurans c
|
|
|
| 27 | c3v7qB_ |
|
not modelled |
73.0 |
12 |
PDB header:rna binding protein
Chain: B: PDB Molecule:probable ribosomal protein ylxq;
PDBTitle: crystal structure of b. subtilis ylxq at 1.55 a resolution
|
|
|
| 28 | d1t0kb_ |
|
not modelled |
71.0 |
21 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 29 | c3o85A_ |
|
not modelled |
67.4 |
11 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:ribosomal protein l7ae;
PDBTitle: giardia lamblia 15.5kd rna binding protein
|
|
|
| 30 | c2zkr6_ |
|
not modelled |
64.9 |
13 |
PDB header:ribosomal protein/rna
Chain: 6: PDB Molecule:60s ribosomal protein l30e;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
|
|
|
| 31 | c3zf7g_ |
|
not modelled |
64.5 |
17 |
PDB header:ribosome
Chain: G: PDB Molecule:
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 32 | c1dvbA_ |
|
not modelled |
63.5 |
8 |
PDB header:electron transport
Chain: A: PDB Molecule:rubrerythrin;
PDBTitle: rubrerythrin
|
|
|
| 33 | c1yuzB_ |
|
not modelled |
62.7 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nigerythrin;
PDBTitle: partially reduced state of nigerythrin
|
|
|
| 34 | c5ewrA_ |
|
not modelled |
59.5 |
13 |
PDB header:rna binding protein
Chain: A: PDB Molecule:box c/d snornp and u4 snrnp component snu13p;
PDBTitle: c merolae u4 snrnp protein snu13
|
|
|
| 35 | c4a1dG_ |
|
not modelled |
59.4 |
21 |
PDB header:ribosome
Chain: G: PDB Molecule:rpl30;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
|
|
|
| 36 | c3cpqB_ |
|
not modelled |
58.6 |
11 |
PDB header:ribosomal protein
Chain: B: PDB Molecule:50s ribosomal protein l30e;
PDBTitle: crystal structure of l30e a ribosomal protein from2 methanocaldococcus jannaschii dsm2661 (mj1044)
|
|
|
| 37 | d1w41a1 |
|
not modelled |
58.3 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 38 | d2bo1a1 |
|
not modelled |
56.1 |
17 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 39 | c3v7eB_ |
|
not modelled |
56.0 |
20 |
PDB header:ribosomal protein/rna
Chain: B: PDB Molecule:ribosome-associated protein l7ae-like;
PDBTitle: crystal structure of ybxf bound to the sam-i riboswitch aptamer
|
|
|
| 40 | c2hr5B_ |
|
not modelled |
55.1 |
21 |
PDB header:metal binding protein
Chain: B: PDB Molecule:rubrerythrin;
PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
|
|
|
| 41 | c3j21Z_ |
|
not modelled |
53.3 |
13 |
PDB header:ribosome
Chain: Z: PDB Molecule:50s ribosomal protein l30e;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
|
|
|
| 42 | d1rlga_ |
|
not modelled |
52.2 |
21 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 43 | d1vqof1 |
|
not modelled |
49.7 |
17 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 44 | c3j3aM_ |
|
not modelled |
49.0 |
14 |
PDB header:ribosome
Chain: M: PDB Molecule:40s ribosomal protein s12;
PDBTitle: structure of the human 40s ribosomal proteins
|
|
|
| 45 | c3zeyF_ |
|
not modelled |
48.2 |
20 |
PDB header:ribosome
Chain: F: PDB Molecule:40s ribosomal protein s12;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 46 | c5xyiM_ |
|
not modelled |
48.2 |
20 |
PDB header:ribosome
Chain: M: PDB Molecule:ribosomal protein l7ae, putative;
PDBTitle: small subunit of trichomonas vaginalis ribosome
|
|
|
| 47 | d1xbia1 |
|
not modelled |
45.3 |
16 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 48 | d1gz0a2 |
|
not modelled |
45.2 |
20 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:RNA 2'-O ribose methyltransferase substrate binding domain |
|
|
| 49 | d2alea1 |
|
not modelled |
43.5 |
17 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 50 | d1jj2f_ |
|
not modelled |
40.5 |
17 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 51 | c5xxuM_ |
|
not modelled |
40.1 |
24 |
PDB header:ribosome
Chain: M: PDB Molecule:ribosomal protein es12;
PDBTitle: small subunit of toxoplasma gondii ribosome
|
|
|
| 52 | d2ozba1 |
|
not modelled |
37.9 |
15 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 53 | d1gz0f2 |
|
not modelled |
36.8 |
20 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:RNA 2'-O ribose methyltransferase substrate binding domain |
|
|
| 54 | c2xznU_ |
|
not modelled |
35.4 |
17 |
PDB header:ribosome
Chain: U: PDB Molecule:ribosomal protein l7ae containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
|
|
| 55 | d2czwa1 |
|
not modelled |
35.0 |
22 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 56 | d2fc3a1 |
|
not modelled |
33.4 |
15 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 57 | d2hyma2 |
|
not modelled |
30.6 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Fibronectin type III
Family:Fibronectin type III |
|
|
| 58 | c3j38M_ |
|
not modelled |
26.7 |
17 |
PDB header:ribosome
Chain: M: PDB Molecule:40s ribosomal protein s12;
PDBTitle: structure of the d. melanogaster 40s ribosomal proteins
|
|
|
| 59 | c2lbwA_ |
|
not modelled |
26.7 |
13 |
PDB header:rna binding protein
Chain: A: PDB Molecule:h/aca ribonucleoprotein complex subunit 2;
PDBTitle: solution structure of the s. cerevisiae h/aca rnp protein nhp2p-s82w2 mutant
|
|
|
| 60 | c6hmsB_ |
|
not modelled |
25.8 |
17 |
PDB header:replication
Chain: B: PDB Molecule:dna polymerase ii large subunit,dna polymerase ii large
PDBTitle: cryo-em map of dna polymerase d from pyrococcus abyssi in complex with2 dna
|
|
|
| 61 | c4e0uB_ |
|
not modelled |
25.4 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:cyclic dipeptide n-prenyltransferase;
PDBTitle: crystal structure of cdpnpt in complex with thiolodiphosphate and (s)-2 benzodiazependione
|
|
|
| 62 | d2gnra1 |
|
not modelled |
24.6 |
6 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:SSO2064-like |
|
|
| 63 | c1vddC_ |
|
not modelled |
23.3 |
16 |
PDB header:recombination
Chain: C: PDB Molecule:recombination protein recr;
PDBTitle: crystal structure of recombinational repair protein recr
|
|
|
| 64 | c5wb4H_ |
|
not modelled |
22.6 |
5 |
PDB header:transferase
Chain: H: PDB Molecule:n-acetylglucosaminyldiphosphoundecaprenol n-acetyl-beta-d-
PDBTitle: crystal structure of the tara wall teichoic acid glycosyltransferase
|
|
|
| 65 | c3mayE_ |
|
not modelled |
22.6 |
15 |
PDB header:heme-binding protein
Chain: E: PDB Molecule:possible exported protein;
PDBTitle: crystal structure of a secreted mycobacterium tuberculosis heme-2 binding protein
|
|
|
| 66 | c4ld7N_ |
|
not modelled |
22.4 |
10 |
PDB header:transferase
Chain: N: PDB Molecule:dimethylallyl tryptophan synthase;
PDBTitle: crystal structure of anapt from neosartorya fischeri
|
|
|
| 67 | d2aifa1 |
|
not modelled |
22.2 |
17 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
|
|
| 68 | c5ijlA_ |
|
not modelled |
21.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase ii large subunit;
PDBTitle: d-family dna polymerase - dp2 subunit (catalytic subunit)
|
|
|
| 69 | c5zvqA_ |
|
not modelled |
19.4 |
12 |
PDB header:recombination
Chain: A: PDB Molecule:recombination protein recr;
PDBTitle: crystal structure of recombination mediator protein recr
|
|
|
| 70 | d2aa4a2 |
|
not modelled |
16.9 |
18 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 71 | c4u3eA_ |
|
not modelled |
16.6 |
5 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside triphosphate reductase;
PDBTitle: anaerobic ribonucleotide reductase
|
|
|
| 72 | c3ve5D_ |
|
not modelled |
15.2 |
16 |
PDB header:recombination
Chain: D: PDB Molecule:recombination protein recr;
PDBTitle: structure of recombination mediator protein recr16-196 deletion mutant
|
|
|
| 73 | c6ok1B_ |
|
not modelled |
15.1 |
12 |
PDB header:transport protein
Chain: B: PDB Molecule:chsh2(duf35);
PDBTitle: ltp2-chsh2(duf35) aldolase
|
|
|
| 74 | c2aa4B_ |
|
not modelled |
13.9 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:putative n-acetylmannosamine kinase;
PDBTitle: crystal structure of escherichia coli putative n-2 acetylmannosamine kinase, new york structural genomics3 consortium
|
|
|
| 75 | c3u5cM_ |
|
not modelled |
13.7 |
18 |
PDB header:ribosome
Chain: M: PDB Molecule:40s ribosomal protein s12;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome a
|
|
|
| 76 | d2ap1a1 |
|
not modelled |
13.4 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 77 | c4htlA_ |
|
not modelled |
13.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:beta-glucoside kinase;
PDBTitle: lmo2764 protein, a putative n-acetylmannosamine kinase, from listeria2 monocytogenes
|
|
|
| 78 | c5mg5W_ |
|
not modelled |
13.2 |
0 |
PDB header:transferase
Chain: W: PDB Molecule:2,4-diacetylphloroglucinol biosynthesis protein;
PDBTitle: a multi-component acyltransferase phlabc from pseudomonas protegens2 soaked with the monoacetylphloroglucinol (mapg)
|
|
|
| 79 | d1vdda_ |
|
not modelled |
13.0 |
16 |
Fold:Recombination protein RecR
Superfamily:Recombination protein RecR
Family:Recombination protein RecR |
|
|
| 80 | c6et9H_ |
|
not modelled |
12.9 |
10 |
PDB header:transferase
Chain: H: PDB Molecule:pfam duf35;
PDBTitle: structure of the acetoacetyl-coa-thiolase/hmg-coa-synthase complex2 from methanothermococcus thermolithotrophicus at 2.75 a
|
|
|
| 81 | c6mrrA_ |
|
not modelled |
12.1 |
20 |
PDB header:de novo protein
Chain: A: PDB Molecule:foldit1;
PDBTitle: de novo designed protein foldit1
|
|
|
| 82 | c2mxtA_ |
|
not modelled |
12.1 |
20 |
PDB header:splicing
Chain: A: PDB Molecule:heterogeneous nuclear ribonucleoprotein q;
PDBTitle: nmr structure of the acidic domain of syncrip (hnrnpq)
|
|
|
| 83 | c4ijaA_ |
|
not modelled |
11.7 |
9 |
PDB header:protein binding
Chain: A: PDB Molecule:xylr protein;
PDBTitle: structure of s. aureus methicillin resistance factor mecr2
|
|
|
| 84 | d1z05a2 |
|
not modelled |
11.5 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 85 | c5z2vB_ |
|
not modelled |
10.9 |
18 |
PDB header:dna binding protein
Chain: B: PDB Molecule:recombination protein recr;
PDBTitle: crystal structure of recr from pseudomonas aeruginosa pao1
|
|
|
| 86 | d1woqa2 |
|
not modelled |
10.8 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ROK |
|
|
| 87 | c2nykA_ |
|
not modelled |
10.6 |
15 |
PDB header:viral protein
Chain: A: PDB Molecule:m157;
PDBTitle: crystal structure of m157 from mouse cytomegalovirus
|
|
|
| 88 | c5heaA_ |
|
not modelled |
9.3 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:putative glycosyltransferase (galt1);
PDBTitle: cgt structure in hexamer
|
|
|
| 89 | c3o3nB_ |
|
not modelled |
8.9 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:beta-subunit 2-hydroxyacyl-coa dehydratase;
PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
|
|
|
| 90 | c3qocD_ |
|
not modelled |
8.9 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative metallopeptidase;
PDBTitle: crystal structure of n-terminal domain (creatinase/prolidase like2 domain) of putative metallopeptidase from corynebacterium diphtheriae
|
|
|
| 91 | c5f9yB_ |
|
not modelled |
8.5 |
11 |
PDB header:ligase
Chain: B: PDB Molecule:aminoacyl-trna synthetase;
PDBTitle: crystal structure of prolyl-trna synthetase from cryptosporidium2 parvum complexed with l-proline and amp
|
|
|
| 92 | c4db3A_ |
|
not modelled |
8.3 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:n-acetyl-d-glucosamine kinase;
PDBTitle: 1.95 angstrom resolution crystal structure of n-acetyl-d-glucosamine2 kinase from vibrio vulnificus.
|
|
|
| 93 | c3d6xA_ |
|
not modelled |
8.3 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:(3r)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase;
PDBTitle: crystal structure of campylobacter jejuni fabz
|
|
|
| 94 | c3ns5B_ |
|
not modelled |
8.3 |
21 |
PDB header:translation
Chain: B: PDB Molecule:eukaryotic translation initiation factor 3 subunit b;
PDBTitle: crystal structure of the rna recognition motif of yeast eif3b residues2 76-161
|
|
|
| 95 | c2atmA_ |
|
not modelled |
7.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronoglucosaminidase;
PDBTitle: crystal structure of the recombinant allergen ves v 2
|
|
|
| 96 | c2ap1A_ |
|
not modelled |
7.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative regulator protein;
PDBTitle: crystal structure of the putative regulatory protein
|
|
|
| 97 | d2apob1 |
|
not modelled |
7.8 |
9 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
|
|
| 98 | c4ltyD_ |
|
not modelled |
7.8 |
12 |
PDB header:hydrolase
Chain: D: PDB Molecule:exonuclease subunit sbcd;
PDBTitle: crystal structure of e.coli sbcd at 1.8 a resolution
|
|
|
| 99 | c1s1iG_ |
|
not modelled |
7.5 |
12 |
PDB header:ribosome
Chain: G: PDB Molecule:60s ribosomal protein l8-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
|
|
|