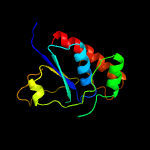
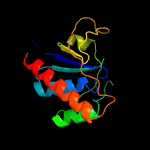
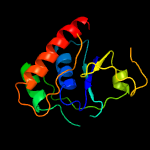
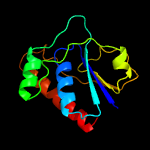
1 c4n7wA_
100.0
21
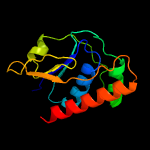
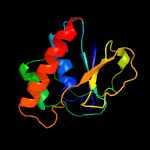
PDB header: transport proteinChain: A: PDB Molecule: transporter, sodium/bile acid symporter family;PDBTitle: crystal structure of the sodium bile acid symporter from yersinia2 frederiksenii
2 c3t38B_
100.0
40
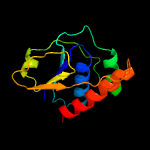
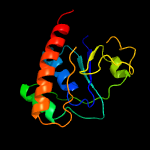
PDB header: oxidoreductaseChain: B: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum thioredoxin-dependent arsenate reductase2 cg_arsc1'
3 d1jf8a_
100.0
34
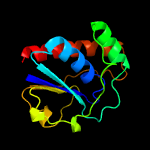
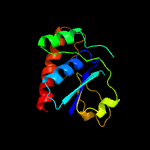
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases
4 c3rh0A_
100.0
34
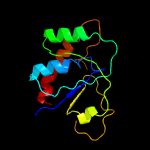
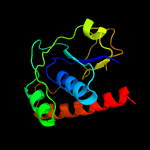
PDB header: oxidoreductaseChain: A: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum mycothiol/mycoredoxin1-dependent arsenate2 reductase cg_arsc2
5 c2l18A_
100.0
37
PDB header: oxidoreductaseChain: A: PDB Molecule: arsenate reductase;PDBTitle: an arsenate reductase in the phosphate binding state
6 d1p8aa_
100.0
22
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases
7 d1y1la_
100.0
31
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases
8 c2fekA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-PDBTitle: structure of a protein tyrosine phosphatase
9 c4lrqC_
100.0
24
PDB header: hydrolaseChain: C: PDB Molecule: phosphotyrosine protein phosphatase;PDBTitle: crystal structure of a low molecular weight phosphotyrosine2 phosphatase from vibrio choleraeo395
10 c4d74A_
100.0
23
PDB header: hydrolaseChain: A: PDB Molecule: protein-tyrosine-phosphatase amsi;PDBTitle: 1.57 a crystal structure of erwinia amylovora tyrosine phosphatase2 amsi
11 c5z3mB_
100.0
26
PDB header: hydrolaseChain: B: PDB Molecule: phosphotyrosine protein phosphatase;PDBTitle: crystal structure of low molecular weight phosphotyrosine phosphatase2 (vclmwptp-2) from vibrio choleraeo395
12 c2wmyH_
100.0
17
PDB header: hydrolaseChain: H: PDB Molecule: putative acid phosphatase wzb;PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
13 c4picA_
100.0
28
PDB header: hydrolaseChain: A: PDB Molecule: arginine phosphatase ywle;PDBTitle: ywle arginine phosphatase from geobacillus stearothermophilus
14 d1dg9a_
100.0
20
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases
15 d5pnta_
100.0
19
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases
16 c2cwdA_
100.0
31
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight phosphotyrosine protein phosphatase;PDBTitle: crystal structure of tt1001 protein from thermus thermophilus hb8
17 c4egsB_
100.0
31
PDB header: isomeraseChain: B: PDB Molecule: ribose 5-phosphate isomerase rpib;PDBTitle: crystal structure analysis of low molecular weight protein tyrosine2 phosphatase from t. tengcongensis
18 d1jl3a_
100.0
34
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases
19 c3jviA_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: protein tyrosine phosphatase;PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
20 c4etiA_
100.0
30
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-phosphatase ywle;PDBTitle: crystal structure of ywle from bacillus subtilis
21 d1d1qa_
not modelled
100.0
18
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases
22 c1zggA_
not modelled
100.0
30
PDB header: hydrolaseChain: A: PDB Molecule: putative low molecular weight protein-tyrosine-PDBTitle: solution structure of a low molecular weight protein2 tyrosine phosphatase from bacillus subtilis
23 c1u2pA_
not modelled
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-PDBTitle: crystal structure of mycobacterium tuberculosis low2 molecular protein tyrosine phosphatase (mptpa) at 1.9a3 resolution
24 c4etmB_
not modelled
100.0
23
PDB header: hydrolaseChain: B: PDB Molecule: low molecular weight protein-tyrosine-phosphatase yfkj;PDBTitle: crystal structure of yfkj from bacillus subtilis
25 c5gotA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight phosphotyrosine phosphatase familyPDBTitle: crystal structure of sp-ptp, low molecular weight protein tyrosine2 phosphatase from streptococcus pyogenes
26 c2gi4A_
not modelled
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: possible phosphotyrosine protein phosphatase;PDBTitle: solution structure of the low molecular weight protein2 tyrosine phosphatase from campylobacter jejuni.
27 c3rofA_
not modelled
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-phosphatase ptpa;PDBTitle: crystal structure of the s. aureus protein tyrosine phosphatase ptpa
28 c5o7bA_
not modelled
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: putative low molecular weight protein-tyrosine-phosphatasePDBTitle: crystal structure of the slr0328 tyrosine phosphatase wzb from2 synechocystis sp. pcc 6803
29 c4cz8A_
not modelled
97.8
13
PDB header: membrane proteinChain: A: PDB Molecule: na+/h+ antiporter, putative;PDBTitle: structure of the sodium proton antiporter panhap from2 pyrococcus abyssii at ph 8.
30 c4bwzA_
not modelled
97.5
10
PDB header: transport proteinChain: A: PDB Molecule: na(+)/h(+) antiporter;PDBTitle: crystal structure of the sodium proton antiporter, napa
31 c5bz3A_
not modelled
97.5
10
PDB header: transport proteinChain: A: PDB Molecule: na(+)/h(+) antiporter;PDBTitle: crystal structure of sodium proton antiporter napa in outward-facing2 conformation.
32 c4czbB_
not modelled
97.4
10
PDB header: membrane proteinChain: B: PDB Molecule: na(+)/h(+) antiporter 1;PDBTitle: structure of the sodium proton antiporter mjnhap1 from2 methanocaldococcus jannaschii at ph 8.
33 c5lm4A_
not modelled
95.1
12
PDB header: transport proteinChain: A: PDB Molecule: excitatory amino acid transporter 1,neutral amino acidPDBTitle: structure of the thermostalilized eaat1 cryst-ii mutant in complex2 with l-asp and the allosteric inhibitor ucph101
34 c3fdfA_
not modelled
93.4
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: fr253;PDBTitle: crystal structure of the serine phosphatase of rna2 polymerase ii ctd (ssu72 superfamily) from drosophila3 melanogaster. orthorhombic crystal form. northeast4 structural genomics consortium target fr253.
35 c4ky0B_
not modelled
90.7
15
PDB header: transport protein, membrane proteinChain: B: PDB Molecule: proton/glutamate symporter, sdf family;PDBTitle: crystal structure of a substrate-free glutamate transporter homologue2 from thermococcus kodakarensis
36 c1zcdA_
not modelled
90.4
19
PDB header: membrane proteinChain: A: PDB Molecule: na(+)/h(+) antiporter 1;PDBTitle: crystal structure of the na+/h+ antiporter nhaa
37 d2nwwa1
not modelled
90.2
15
Fold: Proton glutamate symport proteinSuperfamily: Proton glutamate symport proteinFamily: Proton glutamate symport protein
38 d1jzta_
not modelled
88.9
13
Fold: YjeF N-terminal domain-likeSuperfamily: YjeF N-terminal domain-likeFamily: YjeF N-terminal domain-like
39 c1h6dL_
not modelled
88.8
12
PDB header: protein translocationChain: L: PDB Molecule: precursor form of glucose-fructosePDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
40 c1ofgF_
not modelled
88.4
12
PDB header: oxidoreductaseChain: F: PDB Molecule: glucose-fructose oxidoreductase;PDBTitle: glucose-fructose oxidoreductase
41 c1j6uA_
not modelled
87.4
22
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate-alanine ligase murc;PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase murc (tm0231)2 from thermotoga maritima at 2.3 a resolution
42 c6cauA_
not modelled
87.2
19
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: udp-n-acetylmuramate--alanine ligase from acinetobacter baumannii2 ab5075-uw with amppnp
43 d1vlva2
not modelled
86.1
19
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase
44 c2f00A_
not modelled
84.3
20
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: escherichia coli murc
45 d1j6ua1
not modelled
83.4
22
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
46 d1p3da1
not modelled
80.2
22
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
47 c3o2qB_
not modelled
77.0
36
PDB header: hydrolaseChain: B: PDB Molecule: rna polymerase ii subunit a c-terminal domain phosphatasePDBTitle: crystal structure of the human symplekin-ssu72-ctd phosphopeptide2 complex
48 c3ceaA_
not modelled
76.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: myo-inositol 2-dehydrogenase;PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
49 d1u6ka1
not modelled
75.3
29
Fold: F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)Superfamily: F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)Family: F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
50 c2bibA_
not modelled
74.6
17
PDB header: hydrolaseChain: A: PDB Molecule: teichoic acid phosphorylcholine esterase/ choline bindingPDBTitle: crystal structure of the complete modular teichioic acid2 phosphorylcholine esterase pce (cbpe) from streptococcus pneumoniae
51 d1r8ka_
not modelled
73.9
19
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: PdxA-like
52 c1tvmA_
not modelled
73.4
22
PDB header: transferaseChain: A: PDB Molecule: pts system, galactitol-specific iib component;PDBTitle: nmr structure of enzyme gatb of the galactitol-specific2 phosphoenolpyruvate-dependent phosphotransferase system
53 c1vkrA_
not modelled
73.0
26
PDB header: transferaseChain: A: PDB Molecule: mannitol-specific pts system enzyme iiabc components;PDBTitle: structure of iib domain of the mannitol-specific permease enzyme ii
54 d1vkra_
not modelled
73.0
26
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Lactose/Cellobiose specific IIB subunit
55 d1h6da1
not modelled
72.6
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
56 c5zbyA_
not modelled
71.7
17
PDB header: hydrolaseChain: A: PDB Molecule: hydrogenase maturation protease hyci;PDBTitle: crystal structure of a [nife] hydrogenase maturation protease hyci2 from thermococcus kodakarensis kod1
57 d1duvg2
not modelled
71.2
17
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase
58 c3dfzB_
not modelled
71.2
18
PDB header: oxidoreductaseChain: B: PDB Molecule: precorrin-2 dehydrogenase;PDBTitle: sirc, precorrin-2 dehydrogenase
59 c4ehxA_
not modelled
71.0
18
PDB header: transferaseChain: A: PDB Molecule: tetraacyldisaccharide 4'-kinase;PDBTitle: crystal structure of lpxk from aquifex aeolicus at 1.9 angstrom2 resolution
60 d1dxha2
not modelled
70.9
18
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase
61 d1umdb2
not modelled
69.3
12
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain
62 c4f67A_
not modelled
69.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0176 protein lpg2838;PDBTitle: three dimensional structure of the double mutant of upf0176 protein2 lpg2838 from legionella pneumophila at the resolution 1.8a, northeast3 structural genomics consortium (nesg) target lgr82
63 c3dcjA_
not modelled
67.8
16
PDB header: transferaseChain: A: PDB Molecule: probable 5'-phosphoribosylglycinamide formyltransferasePDBTitle: crystal structure of glycinamide formyltransferase (purn) from2 mycobacterium tuberculosis in complex with 5-methyl-5,6,7,8-3 tetrahydrofolic acid derivative
64 c3hn7A_
not modelled
67.3
16
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate-l-alanine ligase;PDBTitle: crystal structure of a murein peptide ligase mpl (psyc_0032) from2 psychrobacter arcticus 273-4 at 1.65 a resolution
65 c2hi1A_
not modelled
66.2
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 4-hydroxythreonine-4-phosphate dehydrogenase 2;PDBTitle: the structure of a putative 4-hydroxythreonine-4-phosphate2 dehydrogenase from salmonella typhimurium.
66 c3c24A_
not modelled
66.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of a putative oxidoreductase (yp_511008.1) from2 jannaschia sp. ccs1 at 1.62 a resolution
67 c3rbvA_
not modelled
64.7
11
PDB header: sugar binding proteinChain: A: PDB Molecule: sugar 3-ketoreductase;PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
68 c3o2sB_
not modelled
64.2
40
PDB header: hydrolaseChain: B: PDB Molecule: rna polymerase ii subunit a c-terminal domain phosphatasePDBTitle: crystal structure of the human symplekin-ssu72 complex
69 c3eagA_
not modelled
64.1
14
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-PDBTitle: the crystal structure of udp-n-acetylmuramate:l-alanyl-gamma-d-2 glutamyl-meso-diaminopimelate ligase (mpl) from neisseria3 meningitides
70 c6norB_
not modelled
63.6
16
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nad dependent dehydrogenase;PDBTitle: crystal structure of gend2 from gentamicin a biosynthesis in complex2 with nad
71 c4oqyA_
not modelled
62.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: (s)-imine reductase;PDBTitle: streptomyces sp. gf3546 imine reductase
72 d1w85b2
not modelled
61.0
16
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain
73 c2eq8C_
not modelled
59.9
24
PDB header: oxidoreductaseChain: C: PDB Molecule: pyruvate dehydrogenase complex, dihydrolipoamidePDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdp
74 d2jfga1
not modelled
58.8
16
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
75 c5vvwA_
not modelled
58.3
20
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: structure of murc from pseudomonas aeruginosa
76 c4iv5E_
not modelled
58.1
18
PDB header: transferaseChain: E: PDB Molecule: aspartate carbamoyltransferase, putative;PDBTitle: x-ray crystal structure of a putative aspartate carbamoyltransferase2 from trypanosoma cruzi
77 d1fmta2
not modelled
58.0
26
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase
78 c2yvkA_
not modelled
57.5
16
PDB header: isomeraseChain: A: PDB Molecule: methylthioribose-1-phosphate isomerase;PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
79 c3b1fA_
not modelled
56.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: putative prephenate dehydrogenase;PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 mutans
80 c4d3fB_
not modelled
55.7
18
PDB header: oxidoreductaseChain: B: PDB Molecule: imine reductase;PDBTitle: bcsired from bacillus cereus in complex with nadph
81 c4y7jE_
not modelled
55.6
8
PDB header: membrane protein,transport proteinChain: E: PDB Molecule: large conductance mechanosensitive channel protein,PDBTitle: structure of an archaeal mechanosensitive channel in expanded state
82 c4z7fD_
not modelled
54.4
12
PDB header: transport proteinChain: D: PDB Molecule: folate ecf transporter;PDBTitle: crystal structure of folt bound with folic acid
83 c4bucA_
not modelled
53.9
15
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoylalanine--d-glutamate ligase;PDBTitle: crystal structure of murd ligase from thermotoga maritima in apo form
84 c2q62A_
not modelled
53.9
15
PDB header: flavoproteinChain: A: PDB Molecule: arsh;PDBTitle: crystal structure of arsh from sinorhizobium meliloti
85 c2kyrA_
not modelled
53.9
18
PDB header: transferaseChain: A: PDB Molecule: fructose-like phosphotransferase enzyme iib component 1;PDBTitle: solution structure of enzyme iib subunit of pts system from2 escherichia coli k12. northeast structural genomics consortium target3 er315/ontario center for structural proteomics target ec0544
86 c1gqqA_
not modelled
53.0
23
PDB header: cell wall biosynthesisChain: A: PDB Molecule: udp-n-acetylmuramate-l-alanine ligase;PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus influenzae
87 c1e0cA_
not modelled
52.8
17
PDB header: sulfurtransferaseChain: A: PDB Molecule: sulfurtransferase;PDBTitle: sulfurtransferase from azotobacter vinelandii
88 c4o9uB_
not modelled
52.8
18
PDB header: membrane proteinChain: B: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: mechanism of transhydrogenase coupling proton translocation and2 hydride transfer
89 c2bp7F_
not modelled
52.6
20
PDB header: oxidoreductaseChain: F: PDB Molecule: 2-oxoisovalerate dehydrogenase beta subunit;PDBTitle: new crystal form of the pseudomonas putida branched-chain2 dehydrogenase (e1)
90 c2ph5A_
not modelled
52.2
13
PDB header: transferaseChain: A: PDB Molecule: homospermidine synthase;PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
91 c3triB_
not modelled
52.1
19
PDB header: oxidoreductaseChain: B: PDB Molecule: pyrroline-5-carboxylate reductase;PDBTitle: structure of a pyrroline-5-carboxylate reductase (proc) from coxiella2 burnetii
92 c4ldrA_
not modelled
51.4
23
PDB header: isomerase, cell invasionChain: A: PDB Molecule: methylthioribose-1-phosphate isomerase;PDBTitle: structure of the s283y mutant of mrdi
93 c4jqpA_
not modelled
50.9
26
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxythreonine-4-phosphate dehydrogenase;PDBTitle: x-ray crystal structure of a 4-hydroxythreonine-4-phosphate2 dehydrogenase from burkholderia phymatum
94 d1pjqa1
not modelled
50.6
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Siroheme synthase N-terminal domain-like
95 c2eg4B_
not modelled
49.9
23
PDB header: transferaseChain: B: PDB Molecule: probable thiosulfate sulfurtransferase;PDBTitle: crystal structure of probable thiosulfate sulfurtransferase
96 c3db2C_
not modelled
49.9
14
PDB header: oxidoreductaseChain: C: PDB Molecule: putative nadph-dependent oxidoreductase;PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
97 d1qs0b2
not modelled
49.9
18
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain
98 c2moiA_
not modelled
49.8
16
PDB header: membrane proteinChain: A: PDB Molecule: inner membrane protein ygap;PDBTitle: 3d nmr structure of the cytoplasmic rhodanese domain of the inner2 membrane protein ygap from escherichia coli
99 c1pjtB_
not modelled
49.5
19
PDB header: transferase/oxidoreductase/lyaseChain: B: PDB Molecule: siroheme synthase;PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
100 c4ds3A_
not modelled
48.6
10
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 brucella melitensis
101 c4ew6A_
not modelled
47.8
17
PDB header: oxidoreductaseChain: A: PDB Molecule: d-galactose-1-dehydrogenase protein;PDBTitle: crystal structure of d-galactose-1-dehydrogenase protein from2 rhizobium etli
102 c1yxoB_
not modelled
47.7
23
PDB header: oxidoreductaseChain: B: PDB Molecule: 4-hydroxythreonine-4-phosphate dehydrogenase 1;PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein pdxa2 pa0593
103 c2nqqA_
not modelled
46.3
25
PDB header: biosynthetic proteinChain: A: PDB Molecule: molybdopterin biosynthesis protein moea;PDBTitle: moea r137q
104 d1ptma_
not modelled
45.8
19
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: PdxA-like
105 d1t2da1
not modelled
45.5
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
106 c3dzbA_
not modelled
44.5
16
PDB header: biosynthetic proteinChain: A: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 thermophilus
107 c3gt0A_
not modelled
43.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: pyrroline-5-carboxylate reductase;PDBTitle: crystal structure of pyrroline 5-carboxylate reductase from bacillus2 cereus. northeast structural genomics consortium target bcr38b
108 c3ilmD_
not modelled
43.2
11
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: alr3790 protein;PDBTitle: crystal structure of the alr3790 protein from anabaena sp. northeast2 structural genomics consortium target nsr437h
109 d1ryda1
not modelled
42.9
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
110 c5ayvB_
not modelled
42.4
15
PDB header: oxidoreductaseChain: B: PDB Molecule: 2-dehydropantoate 2-reductase;PDBTitle: crystal structure of archaeal ketopantoate reductase complexed with2 coenzyme a and 2-oxopantoate
111 c1gpjA_
not modelled
42.4
21
PDB header: reductaseChain: A: PDB Molecule: glutamyl-trna reductase;PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
112 c3wfjD_
not modelled
41.8
17
PDB header: oxidoreductaseChain: D: PDB Molecule: 2-dehydropantoate 2-reductase;PDBTitle: the complex structure of d-mandelate dehydrogenase with nadh
113 d1rhsa2
not modelled
41.4
8
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)
114 d1f37b_
not modelled
41.1
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioredoxin-like 2Fe-2S ferredoxin
115 c2i99A_
not modelled
41.0
11
PDB header: oxidoreductaseChain: A: PDB Molecule: mu-crystallin homolog;PDBTitle: crystal structure of human mu_crystallin at 2.6 angstrom
116 d2z06a1
not modelled
40.4
14
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: TTHA0625-like
117 c5gziB_
not modelled
39.9
17
PDB header: lyaseChain: B: PDB Molecule: lysine cyclodeaminase;PDBTitle: cyclodeaminase_pa
118 d2fug21
not modelled
39.7
24
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: NQO2-like
119 c5gqsA_
not modelled
39.0
18
PDB header: transport proteinChain: A: PDB Molecule: pts galactitol transporter subunit iib;PDBTitle: nmr based solution structure of pts system, galactitol-specific iib2 component from methicillin resistant staphylococcus aureus
120 c3d1pA_
not modelled
38.6
11
PDB header: transferaseChain: A: PDB Molecule: putative thiosulfate sulfurtransferase yor285w;PDBTitle: atomic resolution structure of uncharacterized protein from2 saccharomyces cerevisiae