| 1 |
|
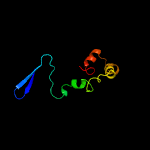
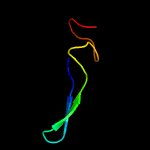
PDB 1ee8 chain A
Region: 24 - 105
Aligned: 69
Modelled: 82
Confidence: 16.3%
Identity: 41%
PDB header:dna binding protein
Chain: A: PDB Molecule:mutm (fpg) protein;
PDBTitle: crystal structure of mutm (fpg) protein from thermus thermophilus hb8
Phyre2
| 2 |
|
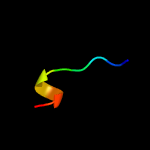
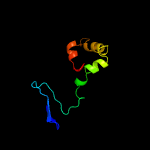
PDB 5zqv chain G
Region: 36 - 45
Aligned: 10
Modelled: 10
Confidence: 14.8%
Identity: 70%
PDB header:hydrolase
Chain: G: PDB Molecule:protein phosphatase 1 regulatory subunit 3a;
PDBTitle: crystal structure of protein phosphate 1 complexed with pp1 binding2 domain of gm
Phyre2
| 3 |
|
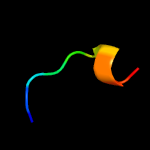
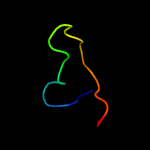
PDB 5zqv chain H
Region: 36 - 45
Aligned: 10
Modelled: 10
Confidence: 14.0%
Identity: 70%
PDB header:hydrolase
Chain: H: PDB Molecule:protein phosphatase 1 regulatory subunit 3a;
PDBTitle: crystal structure of protein phosphate 1 complexed with pp1 binding2 domain of gm
Phyre2
| 4 |
|
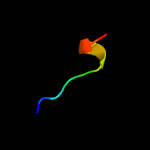
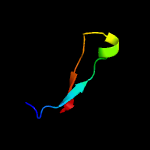
PDB 5zqv chain F
Region: 36 - 45
Aligned: 10
Modelled: 10
Confidence: 13.9%
Identity: 70%
PDB header:hydrolase
Chain: F: PDB Molecule:protein phosphatase 1 regulatory subunit 3a;
PDBTitle: crystal structure of protein phosphate 1 complexed with pp1 binding2 domain of gm
Phyre2
| 5 |
|
PDB 6dno chain B
Region: 36 - 45
Aligned: 10
Modelled: 10
Confidence: 13.7%
Identity: 70%
PDB header:signaling protein
Chain: B: PDB Molecule:protein phosphatase 1 regulatory subunit 3a;
PDBTitle: crystal structure of protein phosphatase 1 (pp1) bound to the muscle2 glycogen-targeting subunit (gm)
Phyre2
| 6 |
|
PDB 3fve chain A
Region: 5 - 54
Aligned: 50
Modelled: 50
Confidence: 12.5%
Identity: 24%
PDB header:isomerase
Chain: A: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of diaminopimelate epimerase mycobacterium2 tuberculosis dapf
Phyre2
| 7 |
|
PDB 5zqv chain E
Region: 36 - 45
Aligned: 10
Modelled: 10
Confidence: 10.7%
Identity: 70%
PDB header:hydrolase
Chain: E: PDB Molecule:protein phosphatase 1 regulatory subunit 3a;
PDBTitle: crystal structure of protein phosphate 1 complexed with pp1 binding2 domain of gm
Phyre2
| 8 |
|
PDB 1l4z chain B
Region: 49 - 82
Aligned: 28
Modelled: 34
Confidence: 9.9%
Identity: 36%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Staphylokinase/streptokinase
Family: Staphylokinase/streptokinase
Phyre2
| 9 |
|
PDB 2b8e chain A domain 1
Region: 33 - 59
Aligned: 26
Modelled: 27
Confidence: 9.7%
Identity: 35%
Fold: HAD-like
Superfamily: HAD-like
Family: Meta-cation ATPase, catalytic domain P
Phyre2
| 10 |
|
PDB 2k87 chain A
Region: 43 - 56
Aligned: 14
Modelled: 14
Confidence: 7.2%
Identity: 21%
PDB header:viral protein, rna binding protein
Chain: A: PDB Molecule:non-structural protein 3 of replicase polyprotein 1a;
PDBTitle: nmr structure of a putative rna binding protein (sars1) from sars2 coronavirus
Phyre2
| 11 |
|
PDB 1t1r chain A domain 3
Region: 3 - 14
Aligned: 12
Modelled: 12
Confidence: 7.2%
Identity: 50%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 12 |
|
PDB 1saz chain A domain 2
Region: 67 - 95
Aligned: 29
Modelled: 29
Confidence: 7.2%
Identity: 31%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Acetokinase-like
Phyre2
| 13 |
|
PDB 3twk chain B
Region: 26 - 105
Aligned: 72
Modelled: 80
Confidence: 7.0%
Identity: 21%
PDB header:hydrolase
Chain: B: PDB Molecule:formamidopyrimidine-dna glycosylase 1;
PDBTitle: crystal structure of arabidopsis thaliana fpg
Phyre2
| 14 |
|
PDB 5xiv chain A
Region: 39 - 48
Aligned: 10
Modelled: 10
Confidence: 6.8%
Identity: 40%
PDB header:unknown function
Chain: A: PDB Molecule:beta-ginkgotide, beta-gb1;
PDBTitle: beta-ginkgotides: hyperdisulfide-constrained peptides from ginkgo2 biloba
Phyre2
| 15 |
|
PDB 5ldw chain C
Region: 42 - 56
Aligned: 15
Modelled: 15
Confidence: 6.8%
Identity: 47%
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,
PDBTitle: structure of mammalian respiratory complex i, class1
Phyre2
| 16 |
|
PDB 6mus chain K
Region: 47 - 69
Aligned: 22
Modelled: 23
Confidence: 6.0%
Identity: 55%
PDB header:rna binding protein/rna
Chain: K: PDB Molecule:uncharacterized protein csm3;
PDBTitle: cryo-em structure of larger csm-crrna-target rna ternary complex in2 type iii-a crispr-cas system
Phyre2
| 17 |
|
PDB 3fhk chain F
Region: 24 - 33
Aligned: 10
Modelled: 10
Confidence: 5.8%
Identity: 60%
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:upf0403 protein yphp;
PDBTitle: crystal structure of apc1446, b.subtilis yphp disulfide isomerase
Phyre2
| 18 |
|
PDB 3opy chain L
Region: 35 - 74
Aligned: 22
Modelled: 25
Confidence: 5.2%
Identity: 59%
PDB header:transferase
Chain: L: PDB Molecule:6-phosphofructo-1-kinase gamma-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
Phyre2
| 19 |
|
PDB 1g6e chain A
Region: 33 - 42
Aligned: 10
Modelled: 10
Confidence: 5.2%
Identity: 60%
Fold: gamma-Crystallin-like
Superfamily: gamma-Crystallin-like
Family: Antifungal protein AFP1
Phyre2
| 20 |
|
PDB 1bu8 chain A domain 2
Region: 36 - 64
Aligned: 28
Modelled: 29
Confidence: 5.1%
Identity: 32%
Fold: alpha/beta-Hydrolases
Superfamily: alpha/beta-Hydrolases
Family: Pancreatic lipase, N-terminal domain
Phyre2
| 21 |
|