| 1 |
|
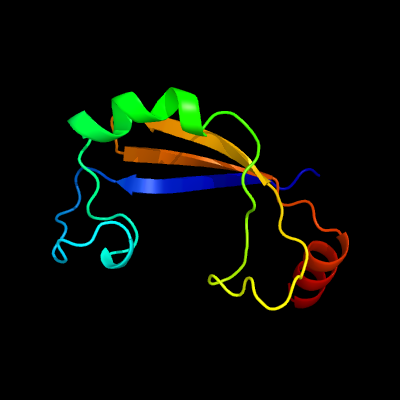
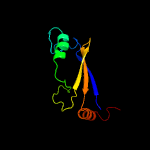
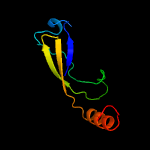
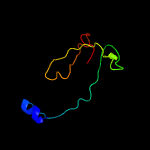
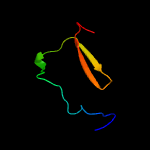
PDB 2pbk chain A domain 1
Region: 15 - 105
Aligned: 82
Modelled: 91
Confidence: 93.9%
Identity: 17%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 2 |
|
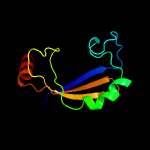
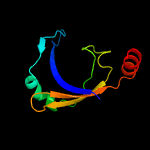
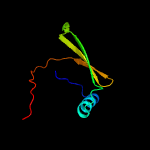
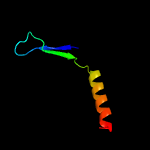
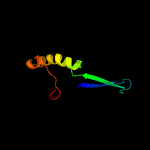
PDB 1o6e chain A
Region: 19 - 93
Aligned: 66
Modelled: 75
Confidence: 93.1%
Identity: 23%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 3 |
|
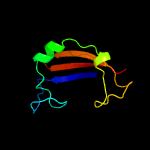
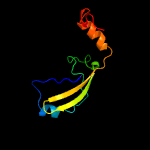
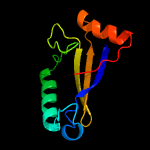
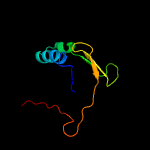
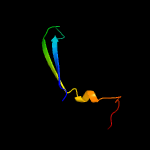
PDB 1ieg chain A
Region: 19 - 105
Aligned: 74
Modelled: 87
Confidence: 92.7%
Identity: 16%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 4 |
|
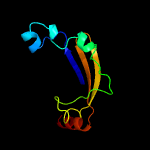
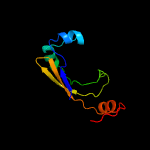
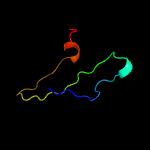
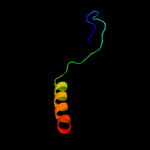
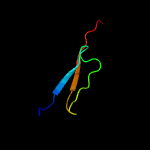
PDB 1vzv chain A
Region: 15 - 112
Aligned: 86
Modelled: 98
Confidence: 90.3%
Identity: 17%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 5 |
|
PDB 3njq chain B
Region: 22 - 105
Aligned: 78
Modelled: 84
Confidence: 88.5%
Identity: 17%
PDB header:viral protein/inhibitor
Chain: B: PDB Molecule:orf 17;
PDBTitle: crystal structure of kaposi's sarcoma-associated herpesvirus protease2 in complex with dimer disruptor
Phyre2
| 6 |
|
PDB 4cx8 chain B
Region: 19 - 114
Aligned: 78
Modelled: 81
Confidence: 88.1%
Identity: 14%
PDB header:viral protein
Chain: B: PDB Molecule:pseudorabies virus protease;
PDBTitle: monomeric pseudorabies virus protease pul26n at 2.5 a resolution
Phyre2
| 7 |
|
PDB 1at3 chain A
Region: 22 - 110
Aligned: 83
Modelled: 89
Confidence: 86.7%
Identity: 18%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 8 |
|
PDB 1jq6 chain A
Region: 19 - 112
Aligned: 71
Modelled: 79
Confidence: 64.3%
Identity: 18%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 9 |
|
PDB 4oy2 chain D
Region: 88 - 159
Aligned: 63
Modelled: 68
Confidence: 56.1%
Identity: 11%
PDB header:transcription
Chain: D: PDB Molecule:transcription initiation factor tfiid subunit 7;
PDBTitle: crystal structure of taf1-taf7, a tfiid subcomplex
Phyre2
| 10 |
|
PDB 5jbl chain A
Region: 16 - 114
Aligned: 95
Modelled: 99
Confidence: 51.8%
Identity: 14%
PDB header:hydrolase
Chain: A: PDB Molecule:prohead core protein protease;
PDBTitle: structure of the bacteriophage t4 capsid assembly protease, gp21.
Phyre2
| 11 |
|
PDB 2ed6 chain A domain 1
Region: 1 - 46
Aligned: 43
Modelled: 46
Confidence: 44.6%
Identity: 16%
Fold: WSSV envelope protein-like
Superfamily: WSSV envelope protein-like
Family: WSSV envelope protein-like
Phyre2
| 12 |
|
PDB 4x8r chain B
Region: 48 - 113
Aligned: 64
Modelled: 66
Confidence: 11.2%
Identity: 13%
PDB header:solute-binding protein
Chain: B: PDB Molecule:trap dicarboxylate transporter, dctp subunit;
PDBTitle: crystal structure of a trap periplasmic solute binding protein from2 rhodobacter sphaeroides (rsph17029_2138, target efi-510205) with3 bound glucuronate
Phyre2
| 13 |
|
PDB 5ur2 chain C
Region: 138 - 177
Aligned: 40
Modelled: 40
Confidence: 10.6%
Identity: 10%
PDB header:oxidoreductase
Chain: C: PDB Molecule:bifunctional protein puta;
PDBTitle: crystal structure of proline utilization a (puta) from bdellovibrio2 bacteriovorus inactivated by n-propargylglycine
Phyre2
| 14 |
|
PDB 4rgw chain B
Region: 88 - 159
Aligned: 65
Modelled: 72
Confidence: 8.5%
Identity: 15%
PDB header:transferase/transcription
Chain: B: PDB Molecule:transcription initiation factor tfiid subunit 7;
PDBTitle: crystal structure of a taf1-taf7 complex in human transcription factor2 iid
Phyre2
| 15 |
|
PDB 4idm chain A
Region: 142 - 177
Aligned: 36
Modelled: 36
Confidence: 8.2%
Identity: 19%
PDB header:oxidoreductase
Chain: A: PDB Molecule:delta-1-pyrroline-5-carboxylate dehydrogenase;
PDBTitle: crystal structure of the delta-pyrroline-5-carboxylate dehydrogenase2 from mycobacterium tuberculosis
Phyre2
| 16 |
|
PDB 2m4v chain A
Region: 105 - 149
Aligned: 45
Modelled: 45
Confidence: 6.4%
Identity: 16%
PDB header:transcription
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: mycobacterium tuberculosis rna polymerase binding protein a (rbpa) and2 its interactions with sigma factors
Phyre2
| 17 |
|
PDB 2hje chain A domain 1
Region: 1 - 66
Aligned: 55
Modelled: 66
Confidence: 5.5%
Identity: 15%
Fold: Profilin-like
Superfamily: Sensory domain-like
Family: LuxQ-periplasmic domain-like
Phyre2
| 18 |
|
PDB 3cax chain A
Region: 135 - 177
Aligned: 42
Modelled: 43
Confidence: 5.5%
Identity: 12%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pf0695;
PDBTitle: crystal structure of uncharacterized protein pf0695
Phyre2
| 19 |
|
PDB 3m86 chain B
Region: 112 - 145
Aligned: 34
Modelled: 34
Confidence: 5.2%
Identity: 9%
PDB header:protein binding
Chain: B: PDB Molecule:amoebiasin-2;
PDBTitle: crystal structure of the cysteine protease inhibitor, ehicp2, from2 entamoeba histolytica
Phyre2