| 1 |
|
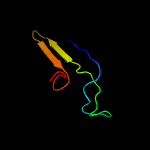
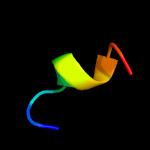
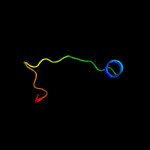
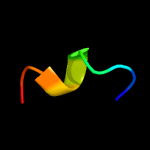
PDB 2co5 chain A domain 1
Region: 6 - 17
Aligned: 12
Modelled: 12
Confidence: 18.3%
Identity: 67%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: F93-like
Phyre2
| 2 |
|
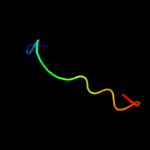
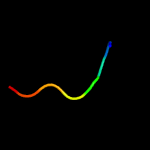
PDB 1z2i chain A
Region: 39 - 85
Aligned: 44
Modelled: 47
Confidence: 14.4%
Identity: 20%
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of agrobacterium tumefaciens malate2 dehydrogenase, new york structural genomics consortium
Phyre2
| 3 |
|
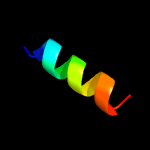
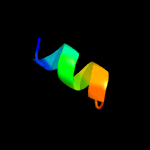
PDB 2lnu chain A
Region: 34 - 85
Aligned: 52
Modelled: 52
Confidence: 13.3%
Identity: 29%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the uncharacterized protein from gene locus2 rrnac0354 of haloarcula marismortui, northeast structural genomics3 consortium target hmr11
Phyre2
| 4 |
|
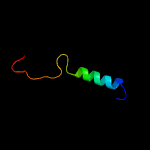
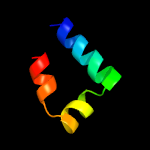
PDB 5m0j chain I
Region: 42 - 55
Aligned: 14
Modelled: 14
Confidence: 11.2%
Identity: 64%
PDB header:rna binding protein
Chain: I: PDB Molecule:swi5-dependent ho expression protein 2,swi5-dependent ho
PDBTitle: crystal structure of the cytoplasmic complex with she2p, she3p, and2 the ash1 mrna e3-localization element
Phyre2
| 5 |
|
PDB 3i0p chain A
Region: 39 - 75
Aligned: 34
Modelled: 37
Confidence: 10.7%
Identity: 26%
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of malate dehydrogenase from entamoeba histolytica
Phyre2
| 6 |
|
PDB 2eyq chain A domain 6
Region: 2 - 15
Aligned: 14
Modelled: 14
Confidence: 10.3%
Identity: 43%
Fold: TRCF domain-like
Superfamily: TRCF domain-like
Family: TRCF domain
Phyre2
| 7 |
|
PDB 2mrb chain A
Region: 75 - 88
Aligned: 14
Modelled: 14
Confidence: 9.1%
Identity: 43%
PDB header:metallothionein
Chain: A: PDB Molecule:cd7 metallothionein-2a;
PDBTitle: three-dimensional structure of rabbit liver cd-72 metallothionein-2a in aqueous solution determined by3 nuclear magnetic resonance
Phyre2
| 8 |
|
PDB 1wtj chain B
Region: 39 - 75
Aligned: 34
Modelled: 37
Confidence: 8.5%
Identity: 32%
PDB header:oxidoreductase
Chain: B: PDB Molecule:ureidoglycolate dehydrogenase;
PDBTitle: crystal structure of delta1-piperideine-2-carboxylate2 reductase from pseudomonas syringae pvar.tomato
Phyre2
| 9 |
|
PDB 1sxm chain A
Region: 81 - 90
Aligned: 10
Modelled: 10
Confidence: 7.4%
Identity: 50%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Scorpion toxin-like
Family: Short-chain scorpion toxins
Phyre2
| 10 |
|
PDB 2l3o chain A
Region: 17 - 50
Aligned: 34
Modelled: 34
Confidence: 7.0%
Identity: 29%
PDB header:cytokine
Chain: A: PDB Molecule:interleukin 3;
PDBTitle: solution structure of murine interleukin 3
Phyre2
| 11 |
|
PDB 1tz5 chain A
Region: 44 - 52
Aligned: 9
Modelled: 9
Confidence: 6.9%
Identity: 56%
PDB header:hormone/growth factor
Chain: A: PDB Molecule:chimera of pancreatic hormone and neuropeptide y;
PDBTitle: [pnpy19-23]-hpp bound to dpc micelles
Phyre2
| 12 |
|
PDB 1mtx chain A
Region: 81 - 90
Aligned: 10
Modelled: 10
Confidence: 6.4%
Identity: 60%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Scorpion toxin-like
Family: Short-chain scorpion toxins
Phyre2
| 13 |
|
PDB 2fyu chain K domain 1
Region: 20 - 40
Aligned: 15
Modelled: 21
Confidence: 6.4%
Identity: 47%
Fold: Single transmembrane helix
Superfamily: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 14 |
|
PDB 2dez chain A
Region: 45 - 52
Aligned: 8
Modelled: 8
Confidence: 6.3%
Identity: 50%
PDB header:neuropeptide
Chain: A: PDB Molecule:peptide yy;
PDBTitle: structure of human pyy
Phyre2
| 15 |
|
PDB 5b52 chain B
Region: 27 - 37
Aligned: 11
Modelled: 11
Confidence: 5.9%
Identity: 64%
PDB header:transcription
Chain: B: PDB Molecule:h-ns family protein mvat;
PDBTitle: crystal structure of the n-terminal domain of h-ns family protein turb
Phyre2
| 16 |
|
PDB 2qsr chain A
Region: 1 - 33
Aligned: 27
Modelled: 33
Confidence: 5.8%
Identity: 30%
PDB header:transcription
Chain: A: PDB Molecule:transcription-repair coupling factor;
PDBTitle: crystal structure of c-terminal domain of transcription-repair2 coupling factor
Phyre2
| 17 |
|
PDB 2bt7 chain A
Region: 14 - 24
Aligned: 11
Modelled: 11
Confidence: 5.8%
Identity: 45%
PDB header:viral protein
Chain: A: PDB Molecule:sigma c;
PDBTitle: structure of the c-terminal receptor-binding domain of avian reovirus2 fibre sigmac, cd crystal form
Phyre2
| 18 |
|
PDB 4ol4 chain A
Region: 32 - 58
Aligned: 27
Modelled: 27
Confidence: 5.5%
Identity: 7%
PDB header:lipid binding protein
Chain: A: PDB Molecule:proline-rich 28 kda antigen;
PDBTitle: crystal structure of secreted proline rich antigen mtc28 (rv0040c)2 from mycobacterium tuberculosis
Phyre2
| 19 |
|
PDB 1hly chain A
Region: 81 - 90
Aligned: 10
Modelled: 10
Confidence: 5.5%
Identity: 60%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Scorpion toxin-like
Family: Short-chain scorpion toxins
Phyre2
| 20 |
|
PDB 1vbi chain A
Region: 39 - 75
Aligned: 34
Modelled: 37
Confidence: 5.5%
Identity: 41%
PDB header:oxidoreductase
Chain: A: PDB Molecule:type 2 malate/lactate dehydrogenase;
PDBTitle: crystal structure of type 2 malate/lactate dehydrogenase from thermus2 thermophilus hb8
Phyre2