| 1 |
|
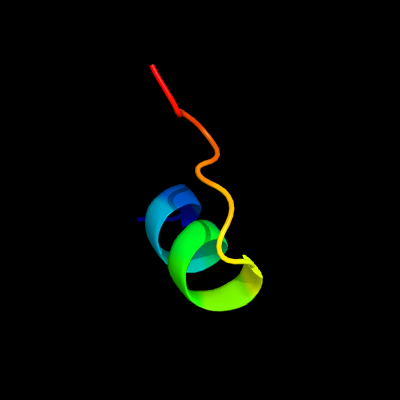
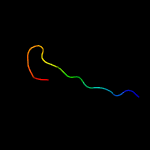
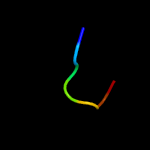
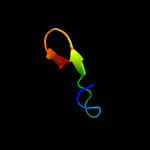
PDB 4dq5 chain B
Region: 7 - 23
Aligned: 17
Modelled: 17
Confidence: 29.4%
Identity: 65%
PDB header:membrane protein
Chain: B: PDB Molecule:membrane protein phi6 p5wt;
PDBTitle: structural investigation of bacteriophage phi6 lysin (wt)
Phyre2
| 2 |
|
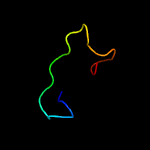
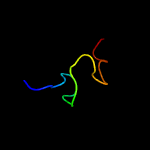
PDB 3ifu chain A
Region: 56 - 80
Aligned: 25
Modelled: 25
Confidence: 26.3%
Identity: 56%
PDB header:transcription
Chain: A: PDB Molecule:non-structural protein;
PDBTitle: the crystal structure of porcine reproductive and respiratory syndrome2 virus (prrsv) leader protease nsp1
Phyre2
| 3 |
|
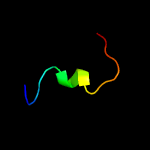
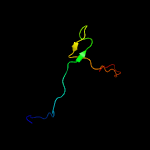
PDB 1ubd chain C domain 1
Region: 19 - 31
Aligned: 13
Modelled: 13
Confidence: 14.3%
Identity: 38%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 4 |
|
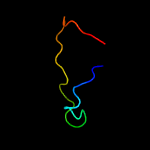
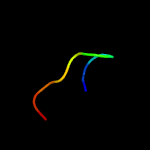
PDB 5mm2 chain B
Region: 75 - 92
Aligned: 18
Modelled: 18
Confidence: 13.1%
Identity: 39%
PDB header:virus
Chain: B: PDB Molecule:capsid protein vp4b;
PDBTitle: nora virus structure
Phyre2
| 5 |
|
PDB 6g4w chain R
Region: 11 - 25
Aligned: 15
Modelled: 15
Confidence: 11.8%
Identity: 47%
PDB header:ribosome
Chain: R: PDB Molecule:40s ribosomal protein s17;
PDBTitle: cryo-em structure of a late human pre-40s ribosomal subunit - state a
Phyre2
| 6 |
|
PDB 5cm2 chain M
Region: 11 - 25
Aligned: 15
Modelled: 15
Confidence: 11.1%
Identity: 33%
PDB header:transferase
Chain: M: PDB Molecule:trna methyltransferase activator subunit;
PDBTitle: insights into molecular plasticity in protein complexes from trm9-2 trm112 trna modifying enzyme crystal structure
Phyre2
| 7 |
|
PDB 2j6a chain A
Region: 11 - 25
Aligned: 15
Modelled: 15
Confidence: 10.7%
Identity: 27%
PDB header:transferase
Chain: A: PDB Molecule:protein trm112;
PDBTitle: crystal structure of s. cerevisiae ynr046w, a zinc finger2 protein from the erf1 methyltransferase complex.
Phyre2
| 8 |
|
PDB 1k28 chain A domain 1
Region: 65 - 92
Aligned: 28
Modelled: 28
Confidence: 8.2%
Identity: 25%
Fold: OB-fold
Superfamily: gp5 N-terminal domain-like
Family: gp4 N-terminal domain-like
Phyre2
| 9 |
|
PDB 1j75 chain A
Region: 83 - 92
Aligned: 10
Modelled: 10
Confidence: 6.6%
Identity: 30%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Z-DNA binding domain
Phyre2
| 10 |
|
PDB 3a44 chain D
Region: 48 - 64
Aligned: 17
Modelled: 17
Confidence: 6.5%
Identity: 18%
PDB header:metal binding protein
Chain: D: PDB Molecule:hydrogenase nickel incorporation protein hypa;
PDBTitle: crystal structure of hypa in the dimeric form
Phyre2
| 11 |
|
PDB 2kdx chain A
Region: 48 - 64
Aligned: 17
Modelled: 17
Confidence: 6.1%
Identity: 18%
PDB header:metal-binding protein
Chain: A: PDB Molecule:hydrogenase/urease nickel incorporation protein
PDBTitle: solution structure of hypa protein
Phyre2
| 12 |
|
PDB 2eln chain A
Region: 18 - 26
Aligned: 9
Modelled: 9
Confidence: 5.9%
Identity: 56%
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 406;
PDBTitle: solution structure of the 11th c2h2 zinc finger of human2 zinc finger protein 406
Phyre2
| 13 |
|
PDB 2l69 chain A
Region: 31 - 49
Aligned: 19
Modelled: 19
Confidence: 5.8%
Identity: 42%
PDB header:de novo protein
Chain: A: PDB Molecule:rossmann 2x3 fold protein;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or28
Phyre2
| 14 |
|
PDB 4xb6 chain D
Region: 49 - 68
Aligned: 20
Modelled: 20
Confidence: 5.5%
Identity: 35%
PDB header:transferase
Chain: D: PDB Molecule:alpha-d-ribose 1-methylphosphonate 5-phosphate c-p lyase;
PDBTitle: structure of the e. coli c-p lyase core complex
Phyre2
| 15 |
|
PDB 2mjc chain A
Region: 54 - 77
Aligned: 24
Modelled: 24
Confidence: 5.5%
Identity: 29%
PDB header:metal binding protein
Chain: A: PDB Molecule:eukaryotic translation initiation factor 3 subunit g;
PDBTitle: zn-binding domain of eukaryotic translation initiation factor 3,2 subunit g
Phyre2
| 16 |
|
PDB 3n40 chain P
Region: 38 - 85
Aligned: 48
Modelled: 48
Confidence: 5.5%
Identity: 25%
PDB header:viral protein
Chain: P: PDB Molecule:p62 envelope glycoprotein;
PDBTitle: crystal structure of the immature envelope glycoprotein complex of2 chikungunya virus.
Phyre2
| 17 |
|
PDB 2lnb chain A
Region: 83 - 92
Aligned: 10
Modelled: 10
Confidence: 5.4%
Identity: 40%
PDB header:immune system
Chain: A: PDB Molecule:z-dna-binding protein 1;
PDBTitle: solution nmr structure of n-terminal domain (6-74) of human zbp12 protein, northeast structural genomics consortium target hr8174a.
Phyre2
| 18 |
|
PDB 4l3u chain A
Region: 28 - 36
Aligned: 9
Modelled: 9
Confidence: 5.3%
Identity: 44%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a duf3571 family protein (abaye3784) from2 acinetobacter baumannii aye at 1.95 a resolution
Phyre2
| 19 |
|
PDB 3hy4 chain A
Region: 75 - 81
Aligned: 7
Modelled: 7
Confidence: 5.2%
Identity: 86%
PDB header:ligase
Chain: A: PDB Molecule:5-formyltetrahydrofolate cyclo-ligase;
PDBTitle: structure of human mthfs with n5-iminium phosphate
Phyre2
| 20 |
|
PDB 1pg5 chain B
Region: 48 - 65
Aligned: 18
Modelled: 18
Confidence: 5.2%
Identity: 28%
PDB header:transferase
Chain: B: PDB Molecule:aspartate carbamoyltransferase regulatory chain;
PDBTitle: crystal structure of the unligated (t-state) aspartate2 transcarbamoylase from the extremely thermophilic archaeon sulfolobus3 acidocaldarius
Phyre2