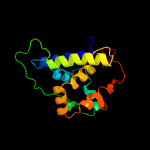
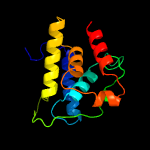
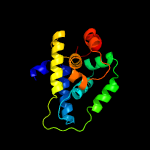
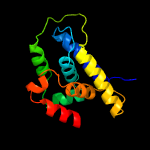
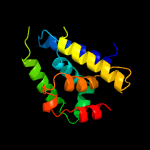
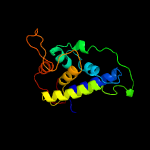
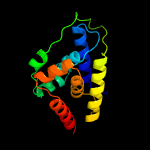
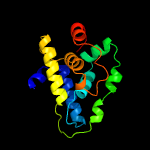
1 c4xbiA_
100.0
22
PDB header: chaperoneChain: A: PDB Molecule: clpb protein, putative,green fluorescent protein;PDBTitle: structure of a malarial protein involved in proteostasis
2 c4y0cB_
100.0
18
PDB header: protein bindingChain: B: PDB Molecule: clp protease-related protein at4g12060, chloroplastic;PDBTitle: the structure of arabidopsis clpt2
3 c3fh2A_
100.0
38
PDB header: hydrolaseChain: A: PDB Molecule: probable atp-dependent protease (heat shock protein);PDBTitle: the crystal structure of the probable atp-dependent protease (heat2 shock protein) from corynebacterium glutamicum
4 c3zriA_
100.0
14
PDB header: chaperoneChain: A: PDB Molecule: clpb protein;PDBTitle: n-domain of clpv from vibrio cholerae
5 c5guiA_
100.0
33
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein clpc1, chloroplastic;PDBTitle: crystal structure of the n-terminal domain of caseinolytic protease2 associated chaperone clpc1 from arabidopsis thaliana
6 c4y0bA_
100.0
17
PDB header: protein bindingChain: A: PDB Molecule: double clp-n motif protein;PDBTitle: the structure of arabidopsis clpt1
7 c2k77A_
100.0
33
PDB header: chaperone, protein bindingChain: A: PDB Molecule: negative regulator of genetic competencePDBTitle: nmr solution structure of the bacillus subtilis clpc n-2 domain
8 c5u2lA_
100.0
23
PDB header: protein bindingChain: A: PDB Molecule: heat shock protein 104;PDBTitle: crystal structure of the hsp104 n-terminal domain from candida2 albicans
9 c5gkmA_
100.0
27
PDB header: chaperoneChain: A: PDB Molecule: at5g51070/k3k7_27;PDBTitle: crystal structure of the n-terminal domain of caseinolytic protease2 associated chaperone clpd from arabidopsis thaliana
10 c4uqwA_
100.0
15
PDB header: chaperoneChain: A: PDB Molecule: protein clpv1;PDBTitle: coevolution of the atpase clpv, the tssb-tssc sheath and2 the accessory hsie protein distinguishes two type vi3 secretion classes
11 c3fesB_
100.0
31
PDB header: atp binding proteinChain: B: PDB Molecule: atp-dependent clp endopeptidase;PDBTitle: crystal structure of the atp-dependent clp protease clpc from2 clostridium difficile
12 d1khya_
100.0
27
Fold: Double Clp-N motifSuperfamily: Double Clp-N motifFamily: Double Clp-N motif
13 d1qvra1
100.0
27
Fold: Double Clp-N motifSuperfamily: Double Clp-N motifFamily: Double Clp-N motif
14 c4irfA_
100.0
19
PDB header: chaperoneChain: A: PDB Molecule: malarial clpb2 atpase/hsp101 protein;PDBTitle: preliminary structural investigations of a malarial protein secretion2 system
15 c4hh5A_
100.0
21
PDB header: protein bindingChain: A: PDB Molecule: putative type vi secretion protein;PDBTitle: n-terminal domain (1-163) of clpv1 atpase from e.coli eaec sci1 t6ss.
16 c6emwX_
99.9
29
PDB header: chaperoneChain: X: PDB Molecule: atp-dependent clp protease atp-binding subunit clpc;PDBTitle: structure of s.aureus clpc in complex with meca
17 d1k6ka_
99.9
19
Fold: Double Clp-N motifSuperfamily: Double Clp-N motifFamily: Double Clp-N motif
18 c3pxgA_
99.9
32
PDB header: protein bindingChain: A: PDB Molecule: negative regulator of genetic competence clpc/mecb;PDBTitle: structure of meca121 and clpc1-485 complex
19 c3pxiB_
99.9
33
PDB header: protein bindingChain: B: PDB Molecule: negative regulator of genetic competence clpc/mecb;PDBTitle: structure of meca108:clpc
20 c1qvrB_
99.9
24
PDB header: chaperoneChain: B: PDB Molecule: clpb protein;PDBTitle: crystal structure analysis of clpb
21 c6azyA_
not modelled
99.9
21
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hsp104;PDBTitle: crystal structure of hsp104 r328m/r757m mutant from calcarisporiella2 thermophila
22 c4d2qC_
not modelled
99.8
26
PDB header: chaperoneChain: C: PDB Molecule: clpb;PDBTitle: negative-stain electron microscopy of e. coli clpb mutant e432a (bap2 form bound to clpp)
23 c5kneD_
not modelled
99.8
22
PDB header: chaperoneChain: D: PDB Molecule: heat shock protein 104;PDBTitle: cryoem reconstruction of hsp104 hexamer
24 c5vy9C_
not modelled
99.7
21
PDB header: chaperoneChain: C: PDB Molecule: heat shock protein 104;PDBTitle: s. cerevisiae hsp104:casein complex, middle domain conformation
25 c6em8F_
not modelled
99.7
32
PDB header: chaperoneChain: F: PDB Molecule: atp-dependent clp protease atp-binding subunit clpc;PDBTitle: s.aureus clpc resting state, c2 symmetrised
26 c6em8E_
not modelled
99.6
33
PDB header: chaperoneChain: E: PDB Molecule: atp-dependent clp protease atp-binding subunit clpc;PDBTitle: s.aureus clpc resting state, c2 symmetrised
27 c1r6bX_
not modelled
99.6
19
PDB header: hydrolaseChain: X: PDB Molecule: clpa protein;PDBTitle: high resolution crystal structure of clpa
28 d1q9ca_
not modelled
86.0
18
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs
29 d1id3c_
not modelled
83.3
32
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
30 d1kx5c_
not modelled
82.4
32
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
31 c2e7xA_
not modelled
81.7
19
PDB header: transcription regulatorChain: A: PDB Molecule: 150aa long hypothetical transcriptional regulator;PDBTitle: structure of the lrp/asnc like transcriptional regulator from2 sulfolobus tokodaii 7 complexed with its cognate ligand
32 d1eqza_
not modelled
80.7
29
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
33 c4pcqC_
not modelled
80.3
19
PDB header: transcriptionChain: C: PDB Molecule: possible transcriptional regulatory protein (probablyPDBTitle: crystal structure of mtbaldr (rv2779c)
34 c3i4pA_
not modelled
79.4
20
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, asnc family;PDBTitle: crystal structure of asnc family transcriptional regulator from2 agrobacterium tumefaciens
35 c2f8nK_
not modelled
78.6
29
PDB header: structural protein/dnaChain: K: PDB Molecule: histone h2a type 1;PDBTitle: 2.9 angstrom x-ray structure of hybrid macroh2a nucleosomes
36 d1s7oa_
not modelled
78.5
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like
37 c1i1gA_
not modelled
78.2
24
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator lrpa;PDBTitle: crystal structure of the lrp-like transcriptional regulator from the2 archaeon pyrococcus furiosus
38 c2ia0A_
not modelled
78.1
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative hth-type transcriptional regulator pf0864;PDBTitle: transcriptional regulatory protein pf0864 from pyrococcus furiosus a2 member of the asnc family (pf0864)
39 c2cfxD_
not modelled
78.1
15
PDB header: transcriptionChain: D: PDB Molecule: hth-type transcriptional regulator lrpc;PDBTitle: structure of b.subtilis lrpc
40 c1u78A_
not modelled
76.9
7
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposable element tc3 transposase;PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
41 c2vbzA_
not modelled
76.7
27
PDB header: dna-binding proteinChain: A: PDB Molecule: transcriptional regulatory protein;PDBTitle: feast or famine regulatory protein (rv3291c)from m.2 tuberculosis complexed with l-tryptophan
42 c2dbbA_
not modelled
76.7
16
PDB header: transcriptional regulatorChain: A: PDB Molecule: putative hth-type transcriptional regulator ph0061;PDBTitle: crystal structure of ph0061
43 d1u35c1
not modelled
76.2
26
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
44 d1vkea_
not modelled
76.0
14
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like
45 c2e1cA_
not modelled
76.0
24
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
46 d1aoic_
not modelled
75.8
29
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
47 d1kx3c_
not modelled
75.7
29
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
48 c2cg4B_
not modelled
75.2
17
PDB header: transcriptionChain: B: PDB Molecule: regulatory protein asnc;PDBTitle: structure of e.coli asnc
49 c2p6tH_
not modelled
75.1
8
PDB header: transcriptionChain: H: PDB Molecule: transcriptional regulator, lrp/asnc family;PDBTitle: crystal structure of transcriptional regulator nmb0573 and l-leucine2 complex from neisseria meningitidis
50 d1tzya_
not modelled
73.1
29
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
51 c2qeuA_
not modelled
72.4
15
PDB header: lyaseChain: A: PDB Molecule: putative carboxymuconolactone decarboxylase;PDBTitle: crystal structure of putative carboxymuconolactone decarboxylase2 (yp_555818.1) from burkholderia xenovorans lb400 at 1.65 a resolution
52 c2gqqB_
not modelled
70.7
17
PDB header: transcriptionChain: B: PDB Molecule: leucine-responsive regulatory protein;PDBTitle: crystal structure of e. coli leucine-responsive regulatory protein2 (lrp)
53 d2cwqa1
not modelled
70.4
19
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: TTHA0727-like
54 c2jssA_
not modelled
68.3
32
PDB header: chaperone/nuclear proteinChain: A: PDB Molecule: chimera of histone h2b.1 and histone h2a.z;PDBTitle: nmr structure of chaperone chz1 complexed with histone2 h2a.z-h2b
55 d2jssa1
not modelled
68.1
32
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
56 c2l4aA_
not modelled
66.8
17
PDB header: dna binding proteinChain: A: PDB Molecule: leucine responsive regulatory protein;PDBTitle: nmr structure of the dna-binding domain of e.coli lrp
57 d2cg4a1
not modelled
66.6
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain
58 c1p8cD_
not modelled
66.5
15
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of tm1620 (apc4843) from thermotoga2 maritima
59 d1rp3a2
not modelled
65.8
5
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
60 c4czdA_
not modelled
64.8
15
PDB header: lyaseChain: A: PDB Molecule: putative transcriptional regulator, asnc family;PDBTitle: sirohaem decarboxylase ahba/b - an enzyme with structural homology2 to the lrp/asnc transcription factor family that is part of the3 alternative haem biosynthesis pathway.
61 d1f66c_
not modelled
63.8
32
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
62 c1f66C_
not modelled
63.8
32
PDB header: structural protein/dnaChain: C: PDB Molecule: histone h2a.z;PDBTitle: 2.6 a crystal structure of a nucleosome core particle2 containing the variant histone h2a.z
63 d2huec1
not modelled
63.2
18
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
64 d1xsva_
not modelled
63.0
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like
65 c4czdD_
not modelled
62.1
10
PDB header: lyaseChain: D: PDB Molecule: putative transcriptional regulator, asnc family;PDBTitle: sirohaem decarboxylase ahba/b - an enzyme with structural homology2 to the lrp/asnc transcription factor family that is part of the3 alternative haem biosynthesis pathway.
66 d1hiod_
not modelled
60.9
17
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
67 c3t72o_
not modelled
59.3
20
PDB header: transcription/dnaChain: O: PDB Molecule: pho box dna (strand 1);PDBTitle: phob(e)-sigma70(4)-(rnap-betha-flap-tip-helix)-dna transcription2 activation sub-complex
68 d1ku3a_
not modelled
58.6
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
69 d2p7vb1
not modelled
57.6
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
70 c1or7A_
not modelled
55.4
23
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma-e factor;PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
71 c6c05F_
not modelled
54.8
25
PDB header: transcriptionChain: F: PDB Molecule: rna polymerase sigma factor siga;PDBTitle: mycobacterium tuberculosis rnap holo/rbpa in relaxed state
72 d1ku7a_
not modelled
54.8
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
73 d1ttya_
not modelled
53.7
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
74 c6in7B_
not modelled
53.7
29
PDB header: transcriptionChain: B: PDB Molecule: rna polymerase sigma-h factor;PDBTitle: crystal structure of algu in complex with muca(cyto)
75 c3vepA_
not modelled
52.2
24
PDB header: membrane protein/transcriptionChain: A: PDB Molecule: probable rna polymerase sigma-d factor;PDBTitle: crystal structure of sigd4 in complex with its negative regulator rsda
76 c5zx3F_
not modelled
51.8
19
PDB header: transcriptionChain: F: PDB Molecule: ecf rna polymerase sigma factor sigh;PDBTitle: mycobacterium tuberculosis rna polymerase holoenzyme with ecf sigma2 factor sigma h
77 d2gena1
not modelled
51.3
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain
78 c1vz0B_
not modelled
51.1
13
PDB header: nuclear proteinChain: B: PDB Molecule: chromosome-partitioning protein spo0j;PDBTitle: chromosome segregation protein spo0j from thermus thermophilus
79 d1id3b_
not modelled
51.0
6
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
80 d2cfxa1
not modelled
50.1
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain
81 c3rkxA_
not modelled
49.0
25
PDB header: ligaseChain: A: PDB Molecule: biotin-[acetyl-coa-carboxylase] ligase;PDBTitle: structural characterisation of staphylococcus aureus biotin protein2 ligase
82 d1i1ga1
not modelled
47.9
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain
83 c5g49A_
not modelled
47.5
15
PDB header: transcriptionChain: A: PDB Molecule: nuclear transcription factor y subunit b-6;PDBTitle: crystal structure of the arabodopsis thaliana histone-fold dimer l1l2 nf-yc3
84 c2qvwA_
not modelled
44.8
60
PDB header: hydrolaseChain: A: PDB Molecule: glp_546_48378_50642;PDBTitle: structure of giardia dicer refined against twinned data
85 d1or7a1
not modelled
44.7
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
86 c4wzsB_
not modelled
44.5
16
PDB header: transcriptionChain: B: PDB Molecule: tata-binding protein-associated phosphoprotein;PDBTitle: crystal structure of the mot1 n-terminal domain in complex with tbp2 and nc2 bound to a promoter dna fragment
87 d1hioa_
not modelled
44.2
29
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
88 c1l9uH_
not modelled
42.4
20
PDB header: transcriptionChain: H: PDB Molecule: sigma factor siga;PDBTitle: thermus aquaticus rna polymerase holoenzyme at 4 a2 resolution
89 c3hugA_
not modelled
42.3
24
PDB header: transcription/membrane proteinChain: A: PDB Molecule: rna polymerase sigma factor;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
90 c5lj5O_
not modelled
42.2
28
PDB header: splicingChain: O: PDB Molecule: pre-mrna-splicing factor cef1;PDBTitle: overall structure of the yeast spliceosome immediately after2 branching.
91 c6a7uA_
not modelled
41.8
17
PDB header: dna binding proteinChain: A: PDB Molecule: histone h2b type 2-e,histone h2a-bbd type 2/3;PDBTitle: crystal structure of histone h2a.bbd-h2b dimer
92 c2llkA_
not modelled
41.8
16
PDB header: cell cycle, transcriptionChain: A: PDB Molecule: cyclin-d-binding myb-like transcription factor 1;PDBTitle: solution nmr structure of the n-terminal myb-like 1 domain of the2 human cyclin-d-binding transcription factor 1 (hdmp1), northeast3 structural genomics consortium (nesg) target id hr8011a
93 d1n1jb_
not modelled
41.6
3
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs
94 c2w7nA_
not modelled
41.4
25
PDB header: transcription/dnaChain: A: PDB Molecule: trfb transcriptional repressor protein;PDBTitle: crystal structure of kora bound to operator dna: insight into2 repressor cooperation in rp4 gene regulation
95 d1v7ba1
not modelled
39.9
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain
96 d2cyya1
not modelled
38.9
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain
97 c5z7iC_
not modelled
38.7
47
PDB header: dna binding protein/dnaChain: C: PDB Molecule: cell cycle regulatory protein gcra;PDBTitle: caulobacter crescentus gcra dna-binding domain(dbd)in complex with2 unmethylated dsdna
98 d1kx5b_
not modelled
38.5
17
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones
99 d2vkea1
not modelled
38.5
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain
100 c5dipB_
not modelled
36.2
13
PDB header: oxidoreductaseChain: B: PDB Molecule: alkyl hydroperoxide reductase ahpd;PDBTitle: crystal structure of lpg0406 in reduced form from legionella2 pneumophila
101 c1zx4B_
not modelled
36.1
21
PDB header: cell cycleChain: B: PDB Molecule: plasmid partition par b protein;PDBTitle: structure of parb bound to dna
102 c2jvlA_
not modelled
35.9
23
PDB header: transcriptionChain: A: PDB Molecule: trmbf1;PDBTitle: nmr structure of the c-terminal domain of mbf1 of trichoderma reesei
103 c5xe7A_
not modelled
35.8
14
PDB header: dna binding proteinChain: A: PDB Molecule: ecf rna polymerase sigma factor sigj;PDBTitle: crystal structure of mycobacterium tuberculosis extracytoplasmic2 function sigma factor sigj
104 c3vwbA_
not modelled
35.6
24
PDB header: transcription/dnaChain: A: PDB Molecule: virulence regulon transcriptional activator virb;PDBTitle: crystal structure of virb core domain (se-met derivative) complexed2 with the cis-acting site (5-bru modifications) upstream icsb promoter
105 c2yusA_
not modelled
34.6
16
PDB header: transcriptionChain: A: PDB Molecule: swi/snf-related matrix-associated actin-PDBTitle: solution structure of the sant domain of human swi/snf-2 related matrix-associated actin-dependent regulator of3 chromatin subfamily c member 1
106 d1gv2a2
not modelled
34.5
32
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain
107 c2ewnA_
not modelled
34.4
20
PDB header: ligase, transcriptionChain: A: PDB Molecule: bira bifunctional protein;PDBTitle: ecoli biotin repressor with co-repressor analog
108 c2kfsA_
not modelled
34.3
11
PDB header: dna-binding proteinChain: A: PDB Molecule: conserved hypothetical regulatory protein;PDBTitle: nmr structure of rv2175c
109 d1smyf2
not modelled
33.6
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
110 c5eybB_
not modelled
33.2
22
PDB header: dna binding protein/dnaChain: B: PDB Molecule: dna-binding protein reb1;PDBTitle: x-ray structure of reb1-ter complex
111 c5y26B_
not modelled
32.9
9
PDB header: dna binding proteinChain: B: PDB Molecule: putative transcription factor c16c4.22;PDBTitle: crystal structure of native dpb4-dpb3
112 d1vi0a1
not modelled
31.2
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain
113 c4ch7A_
not modelled
31.1
16
PDB header: transcriptionChain: A: PDB Molecule: nird-like protein;PDBTitle: crystal structure of the siroheme decarboxylase nirdl
114 c2lf3A_
not modelled
30.6
29
PDB header: signaling proteinChain: A: PDB Molecule: effector protein hopab3;PDBTitle: solution nmr structure of hoppmal_281_385 from pseudomonas syringae2 pv. maculicola str. es4326, midwest center for structural genomics3 target apc40104.5 and northeast structural genomics consortium target4 pst2a
115 d1vkeb_
not modelled
30.1
17
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like
116 c5uxxC_
not modelled
30.1
23
PDB header: dna binding protein/unknown functionChain: C: PDB Molecule: rna polymerase sigma factor;PDBTitle: co-crystal structure of the sigma factor rpoe in complex with the2 anti-sigma factor nepr from bartonella quintana
117 c2lf6A_
not modelled
27.9
26
PDB header: signaling proteinChain: A: PDB Molecule: effector protein hopab1;PDBTitle: solution nmr structure of hopabpph1448_220_320 from pseudomonas2 syringae pv. phaseolicola str. 1448a, midwest center for structural3 genomics target apc40132.4 and northeast structural genomics4 consortium target pst3a
118 c5y26A_
not modelled
26.4
10
PDB header: dna binding proteinChain: A: PDB Molecule: dna polymerase epsilon subunit d;PDBTitle: crystal structure of native dpb4-dpb3
119 c5wurB_
not modelled
26.3
14
PDB header: metal binding proteinChain: B: PDB Molecule: ecf rna polymerase sigma factor sigw;PDBTitle: crystal structure of sigw in complex with its anti-sigma rsiw, an2 oxdized form
120 c6mzlM_
not modelled
25.6
24
PDB header: transcriptionChain: M: PDB Molecule: transcription initiation factor tfiid subunit 9, taf9;PDBTitle: human tfiid canonical state