| 1 |
|
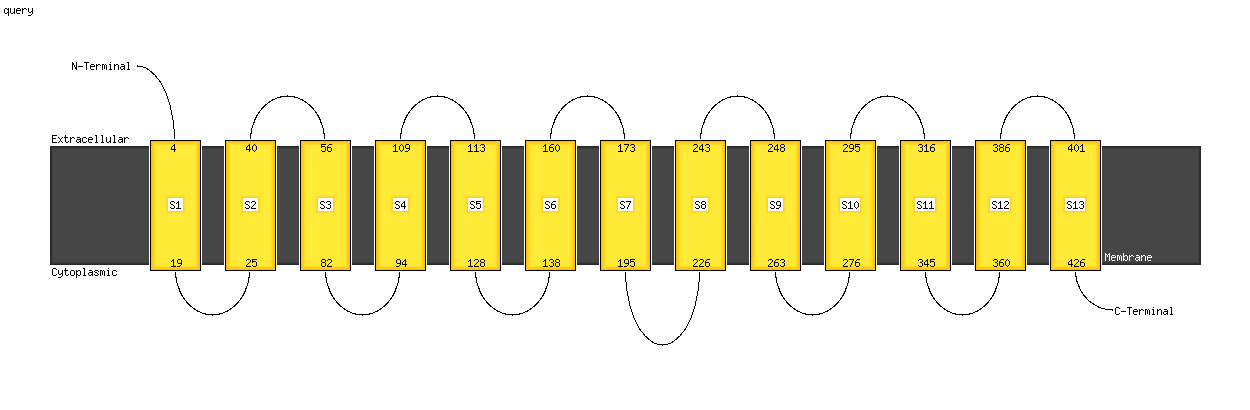
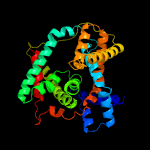
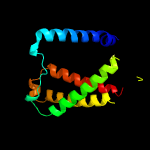
PDB 4f35 chain C
Region: 2 - 424
Aligned: 396
Modelled: 408
Confidence: 100.0%
Identity: 22%
PDB header:transport protein
Chain: C: PDB Molecule:transporter, nadc family;
PDBTitle: crystal structure of a bacterial dicarboxylate/sodium symporter
Phyre2
| 2 |
|
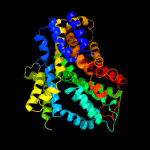
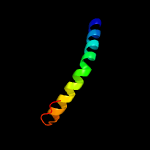
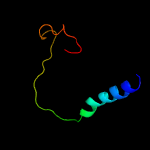
PDB 4r1i chain B
Region: 54 - 427
Aligned: 367
Modelled: 374
Confidence: 100.0%
Identity: 13%
PDB header:membrane protein
Chain: B: PDB Molecule:aminobenzoyl-glutamate transporter;
PDBTitle: structure and function of neisseria gonorrhoeae mtrf illuminates a2 class of antimetabolite efflux pumps
Phyre2
| 3 |
|
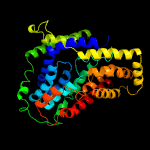
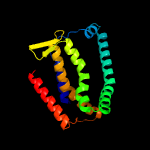
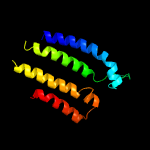
PDB 4r0c chain B
Region: 42 - 426
Aligned: 369
Modelled: 385
Confidence: 99.9%
Identity: 14%
PDB header:membrane protein
Chain: B: PDB Molecule:abgt putative transporter family;
PDBTitle: crystal structure of the alcanivorax borkumensis ydah transporter2 reveals an unusual topology
Phyre2
| 4 |
|
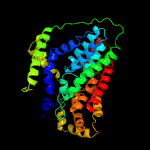
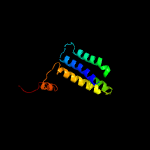
PDB 5a1s chain B
Region: 1 - 331
Aligned: 302
Modelled: 315
Confidence: 93.4%
Identity: 15%
PDB header:transport protein
Chain: B: PDB Molecule:citrate-sodium symporter;
PDBTitle: crystal structure of the sodium-dependent citrate symporter secits2 form salmonella enterica.
Phyre2
| 5 |
|
PDB 3qnq chain D
Region: 178 - 215
Aligned: 38
Modelled: 38
Confidence: 54.8%
Identity: 8%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 6 |
|
PDB 4kpp chain A
Region: 181 - 418
Aligned: 166
Modelled: 179
Confidence: 25.0%
Identity: 17%
PDB header:membrane protein
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of h+/ca2+ exchanger cax
Phyre2
| 7 |
|
PDB 2b6p chain A
Region: 108 - 221
Aligned: 114
Modelled: 114
Confidence: 15.9%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: x-ray structure of lens aquaporin-0 (aqp0) (lens mip) in an open pore2 state
Phyre2
| 8 |
|
PDB 3mk7 chain F
Region: 173 - 240
Aligned: 68
Modelled: 68
Confidence: 8.4%
Identity: 9%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 9 |
|
PDB 6nbx chain G
Region: 166 - 223
Aligned: 58
Modelled: 58
Confidence: 8.2%
Identity: 21%
PDB header:oxidoreductase
Chain: G: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: t.elongatus ndh (data-set 2)
Phyre2
| 10 |
|
PDB 2bbj chain B
Region: 158 - 200
Aligned: 40
Modelled: 43
Confidence: 7.0%
Identity: 20%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 11 |
|
PDB 1fft chain B domain 2
Region: 172 - 238
Aligned: 67
Modelled: 67
Confidence: 6.9%
Identity: 12%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 12 |
|
PDB 1y5i chain C domain 1
Region: 169 - 297
Aligned: 129
Modelled: 129
Confidence: 6.6%
Identity: 9%
Fold: Heme-binding four-helical bundle
Superfamily: Respiratory nitrate reductase 1 gamma chain
Family: Respiratory nitrate reductase 1 gamma chain
Phyre2
| 13 |
|
PDB 4dji chain A
Region: 174 - 224
Aligned: 51
Modelled: 51
Confidence: 6.4%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:probable glutamate/gamma-aminobutyrate antiporter;
PDBTitle: structure of glutamate-gaba antiporter gadc
Phyre2
| 14 |
|
PDB 5lev chain A
Region: 174 - 292
Aligned: 117
Modelled: 119
Confidence: 5.6%
Identity: 12%
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine--dolichyl-phosphate n-
PDBTitle: crystal structure of human udp-n-acetylglucosamine-dolichyl-phosphate2 n-acetylglucosaminephosphotransferase (dpagt1) (v264g mutant)
Phyre2