1 c4g65A_
100.0
25
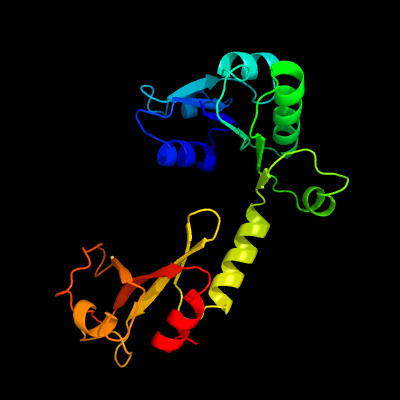
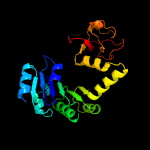
PDB header: transport proteinChain: A: PDB Molecule: trk system potassium uptake protein trka;PDBTitle: potassium transporter peripheral membrane component (trka) from vibrio2 vulnificus
2 c3l4bG_
100.0
27
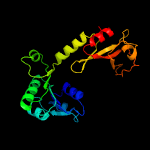
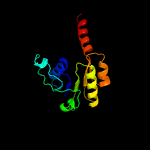
PDB header: transport proteinChain: G: PDB Molecule: trka k+ channel protien tm1088b;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
3 c4gvlB_
100.0
17
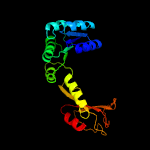
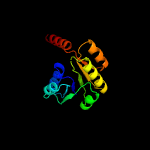
PDB header: transport proteinChain: B: PDB Molecule: trka domain protein;PDBTitle: crystal structure of the gsuk rck domain
4 c4j7cA_
100.0
17
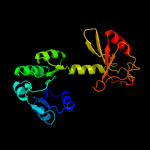
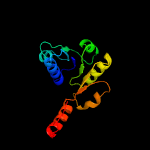
PDB header: transport proteinChain: A: PDB Molecule: ktr system potassium uptake protein a;PDBTitle: ktrab potassium transporter from bacillus subtilis
5 c4gx2B_
100.0
22
PDB header: transport proteinChain: B: PDB Molecule: trka domain protein;PDBTitle: gsuk channel bound to nad
6 c2fy8A_
100.0
19
PDB header: transport proteinChain: A: PDB Molecule: calcium-gated potassium channel mthk;PDBTitle: crystal structure of mthk rck domain in its ligand-free gating-ring2 form
7 c1lnqC_
100.0
19
PDB header: metal transportChain: C: PDB Molecule: potassium channel related protein;PDBTitle: crystal structure of mthk at 3.3 a
8 c3eywA_
99.9
25
PDB header: transport proteinChain: A: PDB Molecule: c-terminal domain of glutathione-regulated potassium-effluxPDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
9 c3fwzA_
99.9
17
PDB header: membrane proteinChain: A: PDB Molecule: inner membrane protein ybal;PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
10 c4hpfB_
99.9
15
PDB header: membrane protein, transport proteinChain: B: PDB Molecule: potassium channel subfamily u member 1;PDBTitle: structure of the human slo3 gating ring
11 d1lssa_
99.9
34
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain
12 c3u6nC_
99.9
16
PDB header: transport proteinChain: C: PDB Molecule: high-conductance ca2+-activated k+ channel protein;PDBTitle: open structure of the bk channel gating ring
13 c4gx5D_
99.9
22
PDB header: transport proteinChain: D: PDB Molecule: trka domain protein;PDBTitle: gsuk channel
14 d2hmva1
99.9
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain
15 d1id1a_
99.9
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain
16 c5nc8B_
99.9
23
PDB header: transport proteinChain: B: PDB Molecule: potassium efflux system protein;PDBTitle: shewanella denitrificans kef ctd in amp bound form
17 c3llvA_
99.9
18
PDB header: nad(p) binding proteinChain: A: PDB Molecule: exopolyphosphatase-related protein;PDBTitle: the crystal structure of the nad(p)-binding domain of an2 exopolyphosphatase-related protein from archaeoglobus fulgidus to3 1.7a
18 c3mt5A_
99.8
15
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: potassium large conductance calcium-activated channel,PDBTitle: crystal structure of the human bk gating apparatus
19 c2g1uA_
99.8
24
PDB header: transport proteinChain: A: PDB Molecule: hypothetical protein tm1088a;PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
20 d2fy8a1
99.8
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain
21 c3c85A_
not modelled
99.8
17
PDB header: transport proteinChain: A: PDB Molecule: putative glutathione-regulated potassium-efflux systemPDBTitle: crystal structure of trka domain of putative glutathione-regulated2 potassium-efflux kefb from vibrio parahaemolyticus
22 c5butG_
not modelled
99.8
22
PDB header: membrane proteinChain: G: PDB Molecule: ktr system potassium uptake protein a,ktr system potassiumPDBTitle: crystal structure of inactive conformation of ktrab k+ transporter
23 c5tj6A_
not modelled
99.7
15
PDB header: membrane proteinChain: A: PDB Molecule: high conductance calcium-activated potassium channel;PDBTitle: ca2+ bound aplysia slo1
24 c5a6eC_
not modelled
99.7
13
PDB header: transportChain: C: PDB Molecule: gating ring of potassium channel subfamily t member 1;PDBTitle: cryo-em structure of the slo2.2 na-activated k channel
25 c5u76A_
not modelled
99.7
11
PDB header: transport proteinChain: A: PDB Molecule: potassium channel subfamily t member 1;PDBTitle: chicken slo2.2 in a closed conformation vitrified in the presence of2 300 mm nacl
26 c3nafA_
not modelled
99.6
16
PDB header: ion transportChain: A: PDB Molecule: calcium-activated potassium channel subunit alpha-1,PDBTitle: structure of the intracellular gating ring from the human high-2 conductance ca2+ gated k+ channel (bk channel)
27 d1e5qa1
not modelled
99.4
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
28 c3jxoB_
not modelled
99.3
30
PDB header: transport proteinChain: B: PDB Molecule: trka-n domain protein;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
29 c4ys2A_
not modelled
99.2
31
PDB header: immune systemChain: A: PDB Molecule: na+/h+ antiporter-like protein;PDBTitle: rck domain with cda
30 d1vcta2
not modelled
99.0
20
Fold: TrkA C-terminal domain-likeSuperfamily: TrkA C-terminal domain-likeFamily: TrkA C-terminal domain-like
31 d2fy8a2
not modelled
99.0
21
Fold: TrkA C-terminal domain-likeSuperfamily: TrkA C-terminal domain-likeFamily: TrkA C-terminal domain-like
32 c2bknA_
not modelled
99.0
19
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical protein ph0236;PDBTitle: structure analysis of unknown function protein
33 c4xttA_
not modelled
98.8
19
PDB header: transport proteinChain: A: PDB Molecule: putative potassium transport protein;PDBTitle: structural studies of potassium transport protein ktra regulator of2 conductance of k+ (rck) c domain in complex with cyclic diadenosine3 monophosphate (c-di-amp)
34 c3ic5A_
not modelled
98.7
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative saccharopine dehydrogenase;PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
35 c5zikC_
not modelled
98.7
15
PDB header: oxidoreductaseChain: C: PDB Molecule: probable 2-dehydropantoate 2-reductase;PDBTitle: crystal structure of ketopantoate reductase from pseudomonas2 aeruginosa
36 d1pjqa1
not modelled
98.7
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Siroheme synthase N-terminal domain-like
37 d2f1ka2
not modelled
98.6
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
38 c2f1kD_
not modelled
98.6
24
PDB header: oxidoreductaseChain: D: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
39 c3wfjD_
not modelled
98.5
19
PDB header: oxidoreductaseChain: D: PDB Molecule: 2-dehydropantoate 2-reductase;PDBTitle: the complex structure of d-mandelate dehydrogenase with nadh
40 c5a9tA_
not modelled
98.5
22
PDB header: oxidoreductaseChain: A: PDB Molecule: putative dehydrogenase;PDBTitle: imine reductase from amycolatopsis orientalis in complex2 with (r)-methyltetrahydroisoquinoline
41 d1jaya_
not modelled
98.4
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
42 c2x4gA_
not modelled
98.4
25
PDB header: isomeraseChain: A: PDB Molecule: nucleoside-diphosphate-sugar epimerase;PDBTitle: crystal structure of pa4631, a nucleoside-diphosphate-sugar epimerase2 from pseudomonas aeruginosa
43 c3qhaB_
not modelled
98.4
15
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of a putative oxidoreductase from mycobacterium2 avium 104
44 c2vrcD_
not modelled
98.4
23
PDB header: oxidoreductaseChain: D: PDB Molecule: triphenylmethane reductase;PDBTitle: crystal structure of the citrobacter sp. triphenylmethane2 reductase complexed with nadp(h)
45 c2ew2B_
not modelled
98.4
22
PDB header: oxidoreductaseChain: B: PDB Molecule: 2-dehydropantoate 2-reductase, putative;PDBTitle: crystal structure of the putative 2-dehydropantoate 2-reductase from2 enterococcus faecalis
46 c3egoB_
not modelled
98.4
17
PDB header: oxidoreductaseChain: B: PDB Molecule: probable 2-dehydropantoate 2-reductase;PDBTitle: crystal structure of probable 2-dehydropantoate 2-reductase pane from2 bacillus subtilis
47 d1dlja2
not modelled
98.4
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
48 c3l6dB_
not modelled
98.4
24
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
49 c2y0dB_
not modelled
98.4
18
PDB header: oxidoreductaseChain: B: PDB Molecule: udp-glucose dehydrogenase;PDBTitle: bcec mutation y10k
50 c4ol9A_
not modelled
98.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: putative 2-dehydropantoate 2-reductase;PDBTitle: crystal structure of putative 2-dehydropantoate 2-reductase pane from2 mycobacterium tuberculosis complexed with nadp and oxamate
51 c2ofpB_
not modelled
98.4
19
PDB header: oxidoreductaseChain: B: PDB Molecule: ketopantoate reductase;PDBTitle: crystal structure of escherichia coli ketopantoate2 reductase in a ternary complex with nadp+ and pantoate
52 c5l3zA_
not modelled
98.4
22
PDB header: oxidoreductaseChain: A: PDB Molecule: polyketide ketoreductase simc7;PDBTitle: polyketide ketoreductase simc7 - binary complex with nadp+
53 c1ks9A_
not modelled
98.4
20
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-dehydropantoate 2-reductase;PDBTitle: ketopantoate reductase from escherichia coli
54 c3gg2B_
not modelled
98.3
18
PDB header: oxidoreductaseChain: B: PDB Molecule: sugar dehydrogenase, udp-glucose/gdp-mannose dehydrogenasePDBTitle: crystal structure of udp-glucose 6-dehydrogenase from porphyromonas2 gingivalis bound to product udp-glucuronate
55 c3c85D_
not modelled
98.3
22
PDB header: transport proteinChain: D: PDB Molecule: putative glutathione-regulated potassium-efflux systemPDBTitle: crystal structure of trka domain of putative glutathione-regulated2 potassium-efflux kefb from vibrio parahaemolyticus
56 c5ayvB_
not modelled
98.3
24
PDB header: oxidoreductaseChain: B: PDB Molecule: 2-dehydropantoate 2-reductase;PDBTitle: crystal structure of archaeal ketopantoate reductase complexed with2 coenzyme a and 2-oxopantoate
57 c1e5lA_
not modelled
98.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine reductase;PDBTitle: apo saccharopine reductase from magnaporthe grisea
58 c3ckyA_
not modelled
98.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-hydroxymethyl glutarate dehydrogenase;PDBTitle: structural and kinetic properties of a beta-hydroxyacid dehydrogenase2 involved in nicotinate fermentation
59 c1bg6A_
not modelled
98.3
13
PDB header: oxidoreductaseChain: A: PDB Molecule: n-(1-d-carboxylethyl)-l-norvaline dehydrogenase;PDBTitle: crystal structure of the n-(1-d-carboxylethyl)-l-norvaline2 dehydrogenase from arthrobacter sp. strain 1c
60 c3dttA_
not modelled
98.3
27
PDB header: oxidoreductaseChain: A: PDB Molecule: nadp oxidoreductase;PDBTitle: crystal structure of a putative f420 dependent nadp-reductase2 (arth_0613) from arthrobacter sp. fb24 at 1.70 a resolution
61 d1ks9a2
not modelled
98.3
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
62 d1pgja2
not modelled
98.3
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
63 c2gf2B_
not modelled
98.3
15
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-hydroxyisobutyrate dehydrogenase;PDBTitle: crystal structure of human hydroxyisobutyrate dehydrogenase
64 c3g0oA_
not modelled
98.3
17
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-hydroxyisobutyrate dehydrogenase;PDBTitle: crystal structure of 3-hydroxyisobutyrate dehydrogenase2 (ygbj) from salmonella typhimurium
65 d2pgda2
not modelled
98.3
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
66 d1mv8a2
not modelled
98.3
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
67 d2c5aa1
not modelled
98.3
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
68 c4wb1B_
not modelled
98.3
21
PDB header: oxidoreductaseChain: B: PDB Molecule: cals8;PDBTitle: crystal structure of cals8 from micromonospora echinospora (p294s2 mutant)
69 c2p5uC_
not modelled
98.3
18
PDB header: isomeraseChain: C: PDB Molecule: udp-glucose 4-epimerase;PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-epimerase2 complex with nad
70 d1pjca1
not modelled
98.3
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
71 c5y8mA_
not modelled
98.3
18
PDB header: oxidoreductaseChain: A: PDB Molecule: probable 3-hydroxyisobutyrate dehydrogenase;PDBTitle: mycobacterium tuberculosis 3-hydroxyisobutyrate dehydrogenase2 (mthibadh) + nad + (r)-3-hydroxyisobutyrate (r-hiba)
72 c4tskA_
not modelled
98.2
19
PDB header: oxidoreductase,isomeraseChain: A: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: ketol-acid reductoisomerase from alicyclobacillus acidocaldarius
73 c4zrmB_
not modelled
98.2
21
PDB header: isomeraseChain: B: PDB Molecule: udp-glucose 4-epimerase;PDBTitle: crystal structure of udp-glucose 4-epimerase (tm0509) from2 hyperthermophilic eubacterium thermotoga maritima
74 c4d3fB_
not modelled
98.2
20
PDB header: oxidoreductaseChain: B: PDB Molecule: imine reductase;PDBTitle: bcsired from bacillus cereus in complex with nadph
75 c3fwnB_
not modelled
98.2
9
PDB header: oxidoreductaseChain: B: PDB Molecule: 6-phosphogluconate dehydrogenase, decarboxylating;PDBTitle: dimeric 6-phosphogluconate dehydrogenase complexed with 6-2 phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate
76 c3ggpA_
not modelled
98.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of prephenate dehydrogenase from a. aeolicus in2 complex with hydroxyphenyl propionate and nad+
77 d1bg6a2
not modelled
98.2
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
78 c4a7pA_
not modelled
98.2
16
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-glucose dehydrogenase;PDBTitle: se-met derivatized ugdg, udp-glucose dehydrogenase from sphingomonas2 elodea
79 c5t57A_
not modelled
98.2
24
PDB header: oxidoreductaseChain: A: PDB Molecule: semialdehyde dehydrogenase nad-binding protein;PDBTitle: crystal structure of a semialdehyde dehydrogenase nad-binding protein2 from cupriavidus necator in complex with calcium and nad
80 c5ojlA_
not modelled
98.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: imine reductase;PDBTitle: imine reductase from aspergillus terreus in complex with nadph4 and2 dibenz[c,e]azepine
81 c2qx7A_
not modelled
98.2
22
PDB header: plant proteinChain: A: PDB Molecule: eugenol synthase 1;PDBTitle: structure of eugenol synthase from ocimum basilicum
82 c3k96B_
not modelled
98.2
19
PDB header: oxidoreductaseChain: B: PDB Molecule: glycerol-3-phosphate dehydrogenase [nad(p)+];PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
83 d1udca_
not modelled
98.2
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
84 c3ktdC_
not modelled
98.2
17
PDB header: oxidoreductaseChain: C: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
85 c1mv8A_
not modelled
98.2
21
PDB header: oxidoreductaseChain: A: PDB Molecule: gdp-mannose 6-dehydrogenase;PDBTitle: 1.55 a crystal structure of a ternary complex of gdp-mannose2 dehydrogenase from psuedomonas aeruginosa
86 c6aqjB_
not modelled
98.2
20
PDB header: isomeraseChain: B: PDB Molecule: ketol-acid reductoisomerase (nadp(+));PDBTitle: crystal structures of staphylococcus aureus ketol-acid2 reductoisomerase in complex with two transition state analogs that3 have biocidal activity.
87 c3plnA_
not modelled
98.2
16
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: crystal structure of klebsiella pneumoniae udp-glucose 6-dehydrogenase2 complexed with udp-glucose
88 c2axqA_
not modelled
98.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
89 c3icpA_
not modelled
98.2
25
PDB header: isomeraseChain: A: PDB Molecule: nad-dependent epimerase/dehydratase;PDBTitle: crystal structure of udp-galactose 4-epimerase
90 c5n2iC_
not modelled
98.2
24
PDB header: oxidoreductaseChain: C: PDB Molecule: reduced coenzyme f420:nadp oxidoreductase;PDBTitle: f420:nadph oxidoreductase from thermobifida fusca with nadp+ bound
91 c5g6sD_
not modelled
98.2
16
PDB header: oxidoreductaseChain: D: PDB Molecule: imine reductase;PDBTitle: imine reductase from aspergillus oryzae in complex with nadp(h) and2 (r)-rasagiline
92 c3vtfA_
not modelled
98.2
20
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: structure of a udp-glucose dehydrogenase from the hyperthermophilic2 archaeon pyrobaculum islandicum
93 c3hwrA_
not modelled
98.2
23
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-dehydropantoate 2-reductase;PDBTitle: crystal structure of pane/apba family ketopantoate reductase2 (yp_299159.1) from ralstonia eutropha jmp134 at 2.15 a resolution
94 c3dojA_
not modelled
98.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: dehydrogenase-like protein;PDBTitle: structure of glyoxylate reductase 1 from arabidopsis (atglyr1)
95 d1txga2
not modelled
98.2
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
96 c2z2vA_
not modelled
98.1
25
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical protein ph1688;PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
97 c1txgA_
not modelled
98.1
27
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerol-3-phosphate dehydrogenase [nad(p)+];PDBTitle: structure of glycerol-3-phosphate dehydrogenase from archaeoglobus2 fulgidus
98 d1n1ea2
not modelled
98.1
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
99 c2zcuA_
not modelled
98.1
24
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized oxidoreductase ytfg;PDBTitle: crystal structure of a new type of nadph-dependent quinone2 oxidoreductase (qor2) from escherichia coli
100 c1i36A_
not modelled
98.1
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein mth1747;PDBTitle: structure of conserved protein mth1747 of unknown function reveals2 structural similarity with 3-hydroxyacid dehydrogenases
101 c3g17H_
not modelled
98.1
23
PDB header: structural genomics, unknown functionChain: H: PDB Molecule: similar to 2-dehydropantoate 2-reductase;PDBTitle: structure of putative 2-dehydropantoate 2-reductase from2 staphylococcus aureus
102 c3d1lB_
not modelled
98.1
16
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nadp oxidoreductase bf3122;PDBTitle: crystal structure of putative nadp oxidoreductase bf3122 from2 bacteroides fragilis
103 c2g5cD_
not modelled
98.1
18
PDB header: oxidoreductaseChain: D: PDB Molecule: prephenate dehydrogenase;PDBTitle: crystal structure of prephenate dehydrogenase from aquifex aeolicus
104 c1dliA_
not modelled
98.1
21
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-glucose dehydrogenase;PDBTitle: the first structure of udp-glucose dehydrogenase (udpgdh) reveals the2 catalytic residues necessary for the two-fold oxidation
105 c1pgjA_
not modelled
98.1
13
PDB header: oxidoreductaseChain: A: PDB Molecule: 6-phosphogluconate dehydrogenase;PDBTitle: x-ray structure of 6-phosphogluconate dehydrogenase from the protozoan2 parasite t. brucei
106 c3c1oA_
not modelled
98.1
22
PDB header: oxidoreductaseChain: A: PDB Molecule: eugenol synthase;PDBTitle: the multiple phenylpropene synthases in both clarkia2 breweri and petunia hybrida represent two distinct lineages
107 c4oqzA_
not modelled
98.1
22
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxidoreductase yfjr;PDBTitle: streptomyces aurantiacus imine reductase
108 c3triB_
not modelled
98.1
21
PDB header: oxidoreductaseChain: B: PDB Molecule: pyrroline-5-carboxylate reductase;PDBTitle: structure of a pyrroline-5-carboxylate reductase (proc) from coxiella2 burnetii
109 c5t8xA_
not modelled
98.1
18
PDB header: oxidoreductaseChain: A: PDB Molecule: prephenate dehydrogenase 1;PDBTitle: prephenate dehydrogenase from soybean
110 c3cumA_
not modelled
98.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: probable 3-hydroxyisobutyrate dehydrogenase;PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
111 d2blla1
not modelled
98.1
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
112 c4pvcB_
not modelled
98.1
17
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-dependent methylglyoxal reductase gre2;PDBTitle: crystal structure of yeast methylglyoxal/ isovaleraldehyde reductase2 gre2
113 c4gbjB_
not modelled
98.1
16
PDB header: oxidoreductaseChain: B: PDB Molecule: 6-phosphogluconate dehydrogenase nad-binding;PDBTitle: crystal structure of nad-binding 6-phosphogluconate dehydrogenase from2 dyadobacter fermentans
114 c3ghyA_
not modelled
98.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: ketopantoate reductase protein;PDBTitle: crystal structure of a putative ketopantoate reductase from ralstonia2 solanacearum molk2
115 c4xdzB_
not modelled
98.0
15
PDB header: oxidoreductaseChain: B: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: holo structure of ketol-acid reductoisomerase from ignisphaera2 aggregans
116 c5ocmA_
not modelled
98.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: nad_gly3p_dh, nad-dependent glycerol-3-phosphatePDBTitle: imine reductase from streptosporangium roseum in complex with nadp+2 and 2,2,2-trifluoroacetophenone hydrate
117 c3gpiA_
not modelled
98.0
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: nad-dependent epimerase/dehydratase;PDBTitle: structure of putative nad-dependent epimerase/dehydratase from2 methylobacillus flagellatus
118 c2pk3B_
not modelled
98.0
25
PDB header: oxidoreductaseChain: B: PDB Molecule: gdp-6-deoxy-d-lyxo-4-hexulose reductase;PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
119 c3dhyC_
not modelled
98.0
21
PDB header: hydrolaseChain: C: PDB Molecule: adenosylhomocysteinase;PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
120 c2uyyD_
not modelled
98.0
24
PDB header: cytokineChain: D: PDB Molecule: n-pac protein;PDBTitle: structure of the cytokine-like nuclear factor n-pac