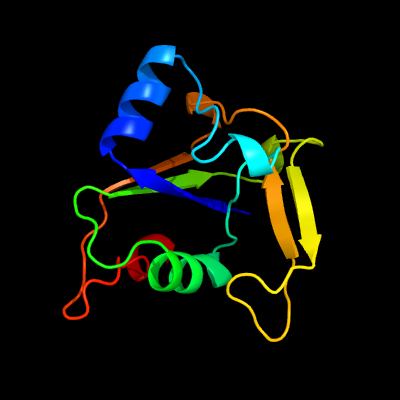
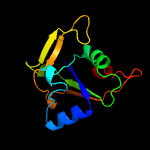
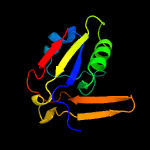
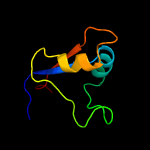
1 d2o0qa1
100.0
31
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: CC0527-like
2 d1wfxa_
86.0
15
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: Tpt1/KptA
3 c6edeA_
74.8
23
PDB header: transferaseChain: A: PDB Molecule: probable rna 2'-phosphotransferase;PDBTitle: trna 2'-phosphotransferase
4 c1o1aP_
28.8
15
PDB header: contractile proteinChain: P: PDB Molecule: skeletal muscle myosin ii;PDBTitle: molecular models of averaged rigor crossbridges from tomograms of2 insect flight muscle
5 d1kl9a2
24.7
40
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
6 c5jknA_
21.7
28
PDB header: hydrolaseChain: A: PDB Molecule: protein fam63a;PDBTitle: crystal structure of deubiquitinase mindy-1
7 d1q46a2
21.4
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
8 d1fnoa3
20.6
13
Fold: Ferredoxin-likeSuperfamily: Bacterial exopeptidase dimerisation domainFamily: Bacterial exopeptidase dimerisation domain
9 d1kjwa2
17.5
9
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases
10 d1wi9a_
17.4
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PCI domain (PINT motif)
11 c2k52A_
17.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mj1198;PDBTitle: structure of uncharacterized protein mj1198 from2 methanocaldococcus jannaschii. northeast structural3 genomics target mjr117b
12 c6hbbA_
16.0
21
PDB header: protein bindingChain: A: PDB Molecule: carbon dioxide concentrating mechanism protein ccmm;PDBTitle: crystal structure of the small subunit-like domain 1 of ccmm from2 synechococcus elongatus (strain pcc 7942)
13 d1ryla_
13.9
18
Fold: Hypothetical protein yfbMSuperfamily: Hypothetical protein yfbMFamily: Hypothetical protein yfbM
14 d1sroa_
13.8
23
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
15 c6k6lA_
13.6
17
PDB header: unknown functionChain: A: PDB Molecule: pseudo deubiquitinase;PDBTitle: ygl082w-catalytic domain
16 c5v77B_
13.0
67
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from neisseria2 gonorrhoeae
17 c1q46A_
12.0
23
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: crystal structure of the eif2 alpha subunit from2 saccharomyces cerevisia
18 c4wsiB_
11.4
15
PDB header: peptide binding proteinChain: B: PDB Molecule: maguk p55 subfamily member 5;PDBTitle: crystal structure of pals1/crb complex
19 c1g8xB_
11.4
17
PDB header: structural proteinChain: B: PDB Molecule: myosin ii heavy chain fused to alpha-actinin 3;PDBTitle: structure of a genetically engineered molecular motor
20 d1r26a_
11.1
9
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase
21 c1yz6A_
not modelled
11.0
20
PDB header: translationChain: A: PDB Molecule: probable translation initiation factor 2 alphaPDBTitle: crystal structure of intact alpha subunit of aif2 from2 pyrococcus abyssi
22 d1s5da_
not modelled
10.9
19
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins
23 d3cx5a2
not modelled
10.6
15
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
24 c4pd3B_
not modelled
10.2
23
PDB header: contractile proteinChain: B: PDB Molecule: nonmuscle myosin heavy chain b, alpha-actinin a chimeraPDBTitle: crystal structure of rigor-like human nonmuscle myosin-2b
25 c5mrgA_
not modelled
10.0
24
PDB header: dna binding proteinChain: A: PDB Molecule: tar dna-binding protein 43;PDBTitle: solution structure of tdp-43 (residues 1-102)
26 d1tr0a_
not modelled
9.5
27
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Plant stress-induced protein
27 c4nnkA_
not modelled
9.5
26
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: structural basis for targeting the ribosomal protein s1 of2 mycobacterium tuberculosis by pyrazinamide
28 d1n0ea_
not modelled
9.3
9
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: Hypothetical protein MraZ
29 c6mr1A_
not modelled
9.3
21
PDB header: protein bindingChain: A: PDB Molecule: carbon dioxide concentrating mechanism protein;PDBTitle: rbcs-like subdomain of ccmm
30 c5i0hA_
not modelled
9.1
15
PDB header: motor proteinChain: A: PDB Molecule: unconventional myosin-x;PDBTitle: crystal structure of myosin x motor domain in pre-powerstroke state
31 d2ahob2
not modelled
8.9
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
32 d1k44a_
not modelled
8.7
14
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK
33 c2ahoB_
not modelled
8.7
19
PDB header: translationChain: B: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: structure of the archaeal initiation factor eif2 alpha-gamma2 heterodimer from sulfolobus solfataricus complexed with gdpnp
34 d3bzka4
not modelled
8.7
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
35 c2k4kA_
not modelled
8.3
26
PDB header: rna binding proteinChain: A: PDB Molecule: general stress protein 13;PDBTitle: solution structure of gsp13 from bacillus subtilis
36 c1n0fF_
not modelled
8.3
9
PDB header: biosynthetic proteinChain: F: PDB Molecule: protein mraz;PDBTitle: crystal structure of a cell division and cell wall2 biosynthesis protein upf0040 from mycoplasma pneumoniae:3 indication of a novel fold with a possible new conserved4 sequence motif
37 c5tbyA_
not modelled
8.3
19
PDB header: contractile proteinChain: A: PDB Molecule: myosin-7;PDBTitle: human beta cardiac heavy meromyosin interacting-heads motif obtained2 by homology modeling (using swiss-model) of human sequence from3 aphonopelma homology model (pdb-3jbh), rigidly fitted to human beta-4 cardiac negatively stained thick filament 3d-reconstruction (emd-5 2240)
38 c2cqoA_
not modelled
8.2
18
PDB header: ribosomeChain: A: PDB Molecule: nucleolar protein of 40 kda;PDBTitle: solution structure of the s1 rna binding domain of human2 hypothetical protein flj11067
39 c6qh2A_
not modelled
7.9
23
PDB header: signaling proteinChain: A: PDB Molecule: polyribonucleotide nucleotidyltransferase;PDBTitle: solution nmr ensemble for a chimeric kh-s1 domain construct of2 exosomal polynucleotide phosphrylase at 298k compiled using the3 comand method
40 c2khiA_
not modelled
7.8
23
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
41 c2n4pA_
not modelled
7.7
24
PDB header: dna binding proteinChain: A: PDB Molecule: tar dna-binding protein 43;PDBTitle: solution structure of the n-terminal domain of tdp-43
42 c3r9lA_
not modelled
7.6
19
PDB header: transferaseChain: A: PDB Molecule: nucleoside diphosphate kinase;PDBTitle: crystal structure of nucleoside diphosphate kinase from giardia2 lamblia featuring a disordered dinucleotide binding site
43 d1zs6a1
not modelled
7.3
23
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK
44 d1wi5a_
not modelled
7.3
11
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
45 d3d85d2
not modelled
7.2
0
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Fibronectin type IIIFamily: Fibronectin type III
46 c3jbhA_
not modelled
7.1
20
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain striated muscle;PDBTitle: two heavy meromyosin interacting-heads motifs flexible docked into2 tarantula thick filament 3d-map allows in depth study of intra- and3 intermolecular interactions
47 d2nn6i2
not modelled
6.9
31
Fold: Barrel-sandwich hybridSuperfamily: Ribosomal L27 protein-likeFamily: ECR1 N-terminal domain-like
48 d1xiqa_
not modelled
6.7
18
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK
49 c3fhkF_
not modelled
6.7
22
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: upf0403 protein yphp;PDBTitle: crystal structure of apc1446, b.subtilis yphp disulfide isomerase
50 d2proc1
not modelled
6.7
36
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Alpha-lytic protease prodomainFamily: Alpha-lytic protease prodomain
51 d1syra_
not modelled
6.6
7
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase
52 c5xq5A_
not modelled
6.6
23
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 5 of the e. coli ribosomal protein s1
53 c1q8kA_
not modelled
6.5
23
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 2PDBTitle: solution structure of alpha subunit of human eif2
54 c2khjA_
not modelled
6.5
20
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
55 c1i84V_
not modelled
6.4
21
PDB header: contractile proteinChain: V: PDB Molecule: smooth muscle myosin heavy chain;PDBTitle: cryo-em structure of the heavy meromyosin subfragment of2 chicken gizzard smooth muscle myosin with regulatory light3 chain in the dephosphorylated state. only c alphas4 provided for regulatory light chain. only backbone atoms5 provided for s2 fragment.
56 c2ycuA_
not modelled
6.3
28
PDB header: motor proteinChain: A: PDB Molecule: non muscle myosin 2c, alpha-actinin;PDBTitle: crystal structure of human non muscle myosin 2c in pre-power stroke2 state
57 d1qled_
not modelled
6.3
20
Fold: Single transmembrane helixSuperfamily: Bacterial aa3 type cytochrome c oxidase subunit IVFamily: Bacterial aa3 type cytochrome c oxidase subunit IV
58 c1kjwA_
not modelled
6.0
9
PDB header: neuropeptideChain: A: PDB Molecule: postsynaptic density protein 95;PDBTitle: sh3-guanylate kinase module from psd-95
59 d1xqia1
not modelled
6.0
14
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK
60 c2mflA_
not modelled
5.9
9
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: domain 2 of e. coli ribosomal protein s1
61 c5i0iB_
not modelled
5.8
16
PDB header: motor proteinChain: B: PDB Molecule: unconventional myosin-x;PDBTitle: crystal structure of myosin x motor domain with 2iq motifs in pre-2 powerstroke state
62 c1br4E_
not modelled
5.7
21
PDB header: muscle proteinChain: E: PDB Molecule: myosin;PDBTitle: smooth muscle myosin motor domain-essential light chain2 complex with mgadp.bef3 bound at the active site
63 c1z6gA_
not modelled
5.7
3
PDB header: transferaseChain: A: PDB Molecule: guanylate kinase;PDBTitle: crystal structure of guanylate kinase from plasmodium falciparum
64 c6n1cB_
not modelled
5.6
27
PDB header: hydrolaseChain: B: PDB Molecule: inorganic pyrophosphatase;PDBTitle: crystal structure of inorganic pyrophosphatase from legionella2 pneumophila philadelphia 1
65 c6igzJ_
not modelled
5.5
30
PDB header: plant proteinChain: J: PDB Molecule: psaj;PDBTitle: structure of psi-lhci
66 d1wkja1
not modelled
5.5
32
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK
67 c3m0zD_
not modelled
5.5
43
PDB header: lyaseChain: D: PDB Molecule: putative aldolase;PDBTitle: crystal structure of putative aldolase from klebsiella pneumoniae.
68 c3dtpA_
not modelled
5.2
22
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth and cardiac muscle;PDBTitle: tarantula heavy meromyosin obtained by flexible docking to tarantula2 muscle thick filament cryo-em 3d-map
69 c4anjA_
not modelled
5.2
15
PDB header: motor protein/metal-bindng proteinChain: A: PDB Molecule: unconventional myosin-vi, green fluorescent protein;PDBTitle: myosin vi (mdinsert2-gfp fusion) pre-powerstroke state (mg.adp.alf4)
70 c3j04A_
not modelled
5.2
21
PDB header: structural proteinChain: A: PDB Molecule: myosin-11;PDBTitle: em structure of the heavy meromyosin subfragment of chick smooth2 muscle myosin with regulatory light chain in phosphorylated state
71 c2ijoA_
not modelled
5.2
27
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: polyprotein;PDBTitle: crystal structure of the west nile virus ns2b-ns3 protease2 complexed with bovine pancreatic trypsin inhibitor
72 d2mysa2
not modelled
5.1
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins
73 c4p7hB_
not modelled
5.1
17
PDB header: motor/fluorescent proteinChain: B: PDB Molecule: myosin-7,green fluorescent protein;PDBTitle: structure of human beta-cardiac myosin motor domain::gfp chimera
74 c3e90C_
not modelled
5.0
27
PDB header: hydrolaseChain: C: PDB Molecule: ns2b cofactor;PDBTitle: west nile vi rus ns2b-ns3protease in complexed with2 inhibitor naph-kkr-h