| 1 |
|
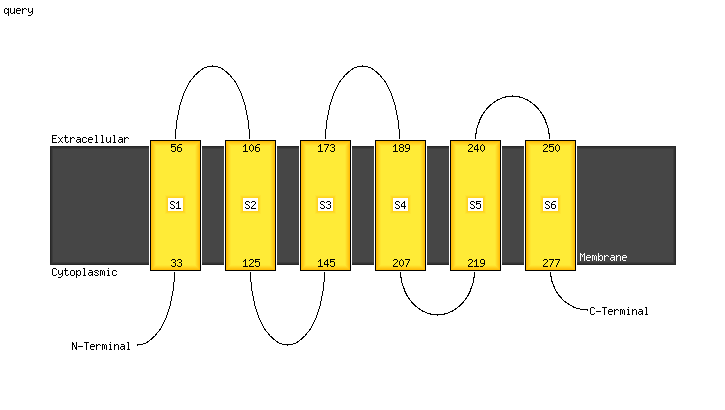
PDB 2r6g chain F domain 1
Region: 190 - 247
Aligned: 56
Modelled: 58
Confidence: 94.1%
Identity: 20%
Fold: MalF N-terminal region-like
Superfamily: MalF N-terminal region-like
Family: MalF N-terminal region-like
Phyre2
| 2 |
|
PDB 6ezn chain H
Region: 129 - 225
Aligned: 92
Modelled: 97
Confidence: 29.7%
Identity: 7%
PDB header:membrane protein
Chain: H: PDB Molecule:dolichyl-diphosphooligosaccharide--protein
PDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
Phyre2
| 3 |
|
PDB 2yn9 chain B
Region: 246 - 266
Aligned: 21
Modelled: 21
Confidence: 10.9%
Identity: 10%
PDB header:hydrolase
Chain: B: PDB Molecule:potassium-transporting atpase subunit beta;
PDBTitle: cryo-em structure of gastric h+,k+-atpase with bound rubidium
Phyre2
| 4 |
|
PDB 2qks chain A
Region: 182 - 270
Aligned: 88
Modelled: 89
Confidence: 10.1%
Identity: 17%
PDB header:metal transport
Chain: A: PDB Molecule:kir3.1-prokaryotic kir channel chimera;
PDBTitle: crystal structure of a kir3.1-prokaryotic kir channel chimera
Phyre2
| 5 |
|
PDB 5lnk chain W
Region: 36 - 59
Aligned: 24
Modelled: 24
Confidence: 9.5%
Identity: 17%
PDB header:oxidoreductase
Chain: W: PDB Molecule:mitochondrial complex i, sgdh subunit;
PDBTitle: entire ovine respiratory complex i
Phyre2
| 6 |
|
PDB 3ech chain C
Region: 3 - 16
Aligned: 14
Modelled: 14
Confidence: 9.1%
Identity: 21%
PDB header:transcription, transcription regulation
Chain: C: PDB Molecule:25-mer fragment of protein armr;
PDBTitle: the marr-family repressor mexr in complex with its antirepressor armr
Phyre2
| 7 |
|
PDB 3b8e chain B
Region: 246 - 265
Aligned: 20
Modelled: 20
Confidence: 8.8%
Identity: 20%
PDB header:hydrolase/transport protein
Chain: B: PDB Molecule:sodium/potassium-transporting atpase subunit
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 8 |
|
PDB 2voy chain B
Region: 2 - 38
Aligned: 36
Modelled: 37
Confidence: 8.2%
Identity: 14%
PDB header:hydrolase
Chain: B: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium atpase 1;
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 9 |
|
PDB 3ixz chain B
Region: 246 - 266
Aligned: 21
Modelled: 21
Confidence: 7.9%
Identity: 10%
PDB header:hydrolase
Chain: B: PDB Molecule:potassium-transporting atpase subunit beta;
PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
Phyre2
| 10 |
|
PDB 2zxe chain B
Region: 246 - 266
Aligned: 21
Modelled: 21
Confidence: 7.5%
Identity: 19%
PDB header:hydrolase/transport protein
Chain: B: PDB Molecule:na+,k+-atpase beta subunit;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
Phyre2
| 11 |
|
PDB 3kdp chain D
Region: 246 - 265
Aligned: 20
Modelled: 20
Confidence: 7.1%
Identity: 20%
PDB header:hydrolase
Chain: D: PDB Molecule:sodium/potassium-transporting atpase subunit beta-1;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 12 |
|
PDB 2xzb chain B
Region: 246 - 266
Aligned: 21
Modelled: 21
Confidence: 6.6%
Identity: 10%
PDB header:hydrolase
Chain: B: PDB Molecule:potassium-transporting atpase subunit beta;
PDBTitle: pig gastric h,k-atpase with bound bef and sch28080
Phyre2
| 13 |
|
PDB 1m9d chain C
Region: 275 - 323
Aligned: 48
Modelled: 49
Confidence: 5.9%
Identity: 17%
Fold: Retrovirus capsid protein, N-terminal core domain
Superfamily: Retrovirus capsid protein, N-terminal core domain
Family: Retrovirus capsid protein, N-terminal core domain
Phyre2
| 14 |
|
PDB 2pxr chain C domain 1
Region: 275 - 323
Aligned: 48
Modelled: 49
Confidence: 5.5%
Identity: 19%
Fold: Retrovirus capsid protein, N-terminal core domain
Superfamily: Retrovirus capsid protein, N-terminal core domain
Family: Retrovirus capsid protein, N-terminal core domain
Phyre2