1 c4qslC_
42.1
17
PDB header: ligaseChain: C: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of listeria monocytogenes pyruvate carboxylase
2 c4qslE_
40.9
17
PDB header: ligaseChain: E: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of listeria monocytogenes pyruvate carboxylase
3 d1jb0f_
22.0
30
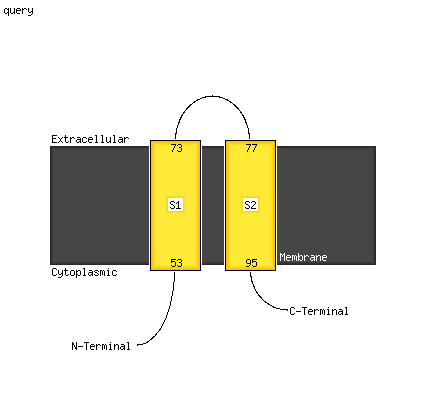
Fold: Single transmembrane helixSuperfamily: Subunit III of photosystem I reaction centre, PsaFFamily: Subunit III of photosystem I reaction centre, PsaF
4 c6fosF_
22.0
35
PDB header: photosynthesisChain: F: PDB Molecule: photosystem i reaction center subunit ii;PDBTitle: cyanidioschyzon merolae photosystem i
5 c3bg3A_
21.7
18
PDB header: ligaseChain: A: PDB Molecule: pyruvate carboxylase, mitochondrial;PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
6 c3bg3B_
17.7
18
PDB header: ligaseChain: B: PDB Molecule: pyruvate carboxylase, mitochondrial;PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
7 c3bg5C_
17.4
17
PDB header: ligaseChain: C: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of staphylococcus aureus pyruvate carboxylase
8 c4kt0F_
17.2
30
PDB header: electron transportChain: F: PDB Molecule: photosystem i subunit iii;PDBTitle: crystal structure of a virus like photosystem i from the2 cyanobacterium synechocystis pcc 6803
9 c6ijjF_
16.3
25
PDB header: membrane proteinChain: F: PDB Molecule: psaf;PDBTitle: photosystem i of chlamydomonas reinhardtii
10 c5ir6A_
15.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: bd-type quinol oxidase subunit i;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
11 c3jcue_
14.5
16
PDB header: membrane proteinChain: E: PDB Molecule: cytochrome b559 subunit alpha;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
12 c2lowA_
14.1
25
PDB header: membrane proteinChain: A: PDB Molecule: apelin receptor;PDBTitle: solution structure of ar55 in 50% hfip
13 d2axte1
14.1
20
Fold: Single transmembrane helixSuperfamily: Cytochrome b559 subunitsFamily: Cytochrome b559 subunits
14 d2r6gf1
13.9
15
Fold: MalF N-terminal region-likeSuperfamily: MalF N-terminal region-likeFamily: MalF N-terminal region-like
15 c3kziE_
13.7
20
PDB header: electron transportChain: E: PDB Molecule: cytochrome b559 subunit alpha;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
16 c5f5oF_
13.6
42
PDB header: nuclear protein/peptideChain: F: PDB Molecule: peptide from polymerase cofactor vp35;PDBTitle: crystal structure of marburg virus nucleoprotein core domain bound to2 vp35 regulation peptide
17 c6hu9p_
13.4
17
PDB header: oxidoreductase/electron transportChain: P: PDB Molecule: cytochrome b-c1 complex subunit rieske, mitochondrial;PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
18 c3rfuC_
13.1
17
PDB header: hydrolase, membrane proteinChain: C: PDB Molecule: copper efflux atpase;PDBTitle: crystal structure of a copper-transporting pib-type atpase
19 d1v54f_
13.1
18
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Cytochrome c oxidase Subunit F
20 c6igzF_
11.9
25
PDB header: plant proteinChain: F: PDB Molecule: psaf;PDBTitle: structure of psi-lhci
21 c4l8nA_
not modelled
11.5
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pdz domain protein;PDBTitle: crystal structure of a pdz domain protein (bdi_1242) from2 parabacteroides distasonis atcc 8503 at 2.50 a resolution
22 c5mkkB_
not modelled
11.3
15
PDB header: transport proteinChain: B: PDB Molecule: multidrug resistance abc transporter atp-binding andPDBTitle: crystal structure of the heterodimeric abc transporter tmrab, a2 homolog of the antigen translocation complex tap
23 c2y69S_
not modelled
11.1
18
PDB header: electron transportChain: S: PDB Molecule: cytochrome c oxidase subunit 5b;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular oxygen
24 c4p6vD_
not modelled
10.5
8
PDB header: oxidoreductaseChain: D: PDB Molecule: na(+)-translocating nadh-quinone reductase subunit d;PDBTitle: crystal structure of the na+-translocating nadh: ubiquinone2 oxidoreductase from vibrio cholerae
25 c4cmxB_
not modelled
10.4
32
PDB header: nuclear proteinChain: B: PDB Molecule: rv3378c;PDBTitle: crystal structure of rv3378c
26 c5mu5A_
not modelled
9.7
26
PDB header: transferaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: structure of maf glycosyltransferase from magnetospirillum magneticum2 amb-1
27 d1ehkb2
not modelled
9.6
20
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region
28 c2kjqA_
not modelled
8.3
27
PDB header: replicationChain: A: PDB Molecule: dnaa-related protein;PDBTitle: solution structure of protein nmb1076 from neisseria meningitidis.2 northeast structural genomics consortium target mr101b.
29 c5vz0D_
not modelled
7.6
20
PDB header: ligaseChain: D: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of lactococcus lactis pyruvate carboxylase g746a2 mutant in complex with cyclic-di-amp
30 c4qvrA_
not modelled
7.5
13
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized hypothetical protein ftt_1539c;PDBTitle: 2.3 angstrom crystal structure of hypothetical protein ftt1539c from2 francisella tularensis.
31 c3dinE_
not modelled
7.3
18
PDB header: membrane protein, protein transportChain: E: PDB Molecule: preprotein translocase subunit secg;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
32 c4aa3A_
not modelled
7.2
13
PDB header: transport proteinChain: A: PDB Molecule: atp-binding cassette sub-family b member 10,PDBTitle: structure of the human mitochondrial abc transporter,2 abcb10 (plate form)
33 c3b5wE_
not modelled
7.0
9
PDB header: membrane proteinChain: E: PDB Molecule: lipid a export atp-binding/permease protein msba;PDBTitle: crystal structure of eschericia coli msba
34 c2k9pA_
not modelled
7.0
13
PDB header: membrane proteinChain: A: PDB Molecule: pheromone alpha factor receptor;PDBTitle: structure of tm1_tm2 in lppg micelles
35 c1z65A_
not modelled
6.9
13
PDB header: unknown functionChain: A: PDB Molecule: prion-like protein doppel;PDBTitle: mouse doppel 1-30 peptide
36 c2m20B_
not modelled
6.4
30
PDB header: signaling proteinChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: egfr transmembrane - juxtamembrane (tm-jm) segment in bicelles: md2 guided nmr refined structure.
37 c2j5dA_
not modelled
6.4
50
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interacting protein 3;PDBTitle: nmr structure of bnip3 transmembrane domain in lipid bicelles
38 d1f46a_
not modelled
6.4
16
Fold: TBP-likeSuperfamily: Cell-division protein ZipA, C-terminal domainFamily: Cell-division protein ZipA, C-terminal domain
39 c1du1A_
not modelled
6.3
27
PDB header: signaling proteinChain: A: PDB Molecule: skeletal dihydropyridine receptor;PDBTitle: peptide fragment thr671-leu690 of the rabbit skeletal2 dihydropyridine receptor
40 c1jzpA_
not modelled
6.3
27
PDB header: signaling proteinChain: A: PDB Molecule: skeletal dihydropydrine receptor;PDBTitle: modified peptide a (d18-a1) of the rabbit skeletal2 dihydropyridine receptor
41 c6go1A_
not modelled
6.1
29
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase-like protein;PDBTitle: crystal structure of a bacillus anthracis peptidoglycan deacetylase
42 c2ypdB_
not modelled
6.1
15
PDB header: oxidoreductaseChain: B: PDB Molecule: probable jmjc domain-containing histone demethylation protPDBTitle: crystal structure of the jumonji domain of human jumonji domain2 containing 1c protein
43 c5fw5C_
not modelled
6.1
67
PDB header: hydrolaseChain: C: PDB Molecule: non-structural protein 3;PDBTitle: crystal structure of human g3bp1 in complex with semliki forest virus2 nsp3-25 comprising two fgdf motives
44 c3g5uB_
not modelled
6.0
19
PDB header: membrane proteinChain: B: PDB Molecule: multidrug resistance protein 1a;PDBTitle: structure of p-glycoprotein reveals a molecular basis for2 poly-specific drug binding
45 d2dk3a1
not modelled
6.0
39
Fold: SH3-like barrelSuperfamily: Mib/herc2 domain-likeFamily: Mib/herc2 domain
46 c5b2gG_
not modelled
5.8
42
PDB header: membrane proteinChain: G: PDB Molecule: endolysin,claudin-4;PDBTitle: crystal structure of human claudin-4 in complex with c-terminal2 fragment of clostridium perfringens enterotoxin
47 c3dkmA_
not modelled
5.8
39
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase hectd1;PDBTitle: crystal structure of the hectd1 cph domain
48 c3qdrB_
not modelled
5.5
38
PDB header: protein transport/toxinChain: B: PDB Molecule: colicin-a;PDBTitle: structural characterization of the interaction of colicin a, colicin2 n, and tolb with the tolaiii translocon
49 c3j08A_
not modelled
5.5
13
PDB header: hydrolase, metal transportChain: A: PDB Molecule: copper-exporting p-type atpase a;PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
50 c2kkuA_
not modelled
5.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of protein af2351 from archaeoglobus fulgidus.2 northeast structural genomics consortium target att9/ontario center3 for structural proteomics target af2351
51 c5aynA_
not modelled
5.2
8
PDB header: transport proteinChain: A: PDB Molecule: solute carrier family 39 (iron-regulated transporter);PDBTitle: crystal structure of a bacterial homologue of iron transporter2 ferroportin in outward-facing state