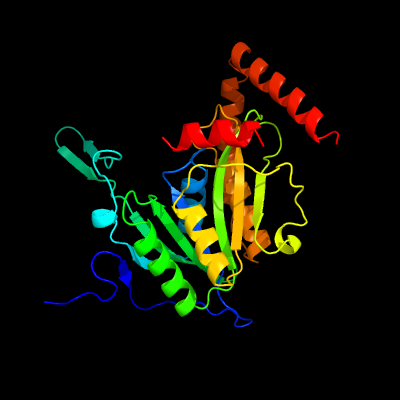
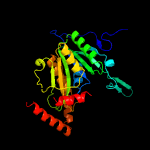
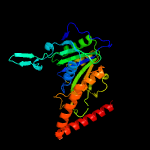
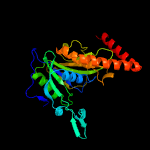
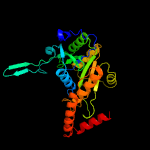
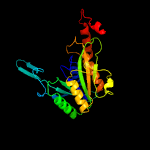
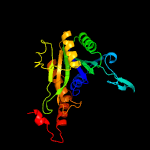
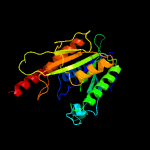
1 c2wamB_
100.0
99
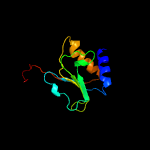
PDB header: unknown functionChain: B: PDB Molecule: conserved hypothetical alanine and leucine richPDBTitle: crystal structure of mycobacterium tuberculosis unknown2 function protein rv2714
2 c2p90B_
100.0
47
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein cgl1923;PDBTitle: the crystal structure of a protein of unknown function from2 corynebacterium glutamicum atcc 13032
3 d2p90a1
100.0
45
Fold: Phosphorylase/hydrolase-likeSuperfamily: Cgl1923-likeFamily: Cgl1923-like
4 c3e35A_
100.0
40
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein sco1997;PDBTitle: actinobacteria-specific protein of unknown function, sco1997
5 c5un02_
100.0
26
PDB header: apoptosisChain: 2: PDB Molecule: proteasome assembly chaperone 2 (pac2) homologue rv2125;PDB Fragment: unp residues 17-260;
PDBTitle: crystal structure of mycobacterium tuberculosis proteasome-assembly2 chaperone homologue rv2125
6 c3mnfA_
100.0
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pac2 family protein;PDBTitle: crystal structure of pac2 family protein from streptomyces avermitilis2 ma
7 c3vr0D_
100.0
18
PDB header: protein bindingChain: D: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of pyrococcus furiosus pbab, an archaeal proteasome2 activator
8 c3gaaB_
100.0
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ta1441;PDBTitle: the crystal structure of the protein with unknown function from2 thermoplasma acidophilum
9 c3wz2C_
100.0
13
PDB header: chaperoneChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of pyrococcus furiosus pbaa, an archaeal homolog of2 proteasome-assembly chaperone
10 c4g4sP_
96.9
14
PDB header: hydrolase/chaperoneChain: P: PDB Molecule: proteasome assembly chaperone 2;PDBTitle: structure of proteasome-pba1-pba2 complex
11 c2p6pB_
88.9
11
PDB header: transferaseChain: B: PDB Molecule: glycosyl transferase;PDBTitle: x-ray crystal structure of c-c bond-forming dtdp-d-olivose-transferase2 urdgt2
12 c3khsB_
83.9
17
PDB header: hydrolaseChain: B: PDB Molecule: purine nucleoside phosphorylase;PDBTitle: crystal structure of grouper iridovirus purine nucleoside2 phosphorylase
13 d1b26a1
77.9
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
14 c1bvuF_
77.9
20
PDB header: oxidoreductaseChain: F: PDB Molecule: protein (glutamate dehydrogenase);PDBTitle: glutamate dehydrogenase from thermococcus litoralis
15 d1gtma1
77.2
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
16 c2tmgD_
74.4
26
PDB header: oxidoreductaseChain: D: PDB Molecule: protein (glutamate dehydrogenase);PDBTitle: thermotoga maritima glutamate dehydrogenase mutant s128r,2 t158e, n117r, s160e
17 d1bvua1
72.5
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
18 d1euza1
71.6
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
19 d1b5ta_
70.1
15
Fold: TIM beta/alpha-barrelSuperfamily: FAD-linked oxidoreductaseFamily: Methylenetetrahydrofolate reductase
20 d2dsta1
65.5
30
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: TTHA1544-like
21 c2fmoA_
not modelled
65.2
14
PDB header: oxidoreductaseChain: A: PDB Molecule: 5,10-methylenetetrahydrofolate reductase;PDBTitle: ala177val mutant of e. coli methylenetetrahydrofolate2 reductase
22 d1hwxa1
not modelled
61.8
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
23 c2h04A_
not modelled
58.9
28
PDB header: hydrolaseChain: A: PDB Molecule: protein tyrosine phosphatase, receptor type, b,;PDBTitle: structural studies of protein tyrosine phosphatase beta catalytic2 domain in complex with inhibitors
24 c5jvsA_
not modelled
58.1
23
PDB header: motor proteinChain: A: PDB Molecule: chimera protein of kinesin heavy chain and microtubule-PDBTitle: the neck-linker + dal and alpha 7 helix of drosophila melanogaster2 kinesin-1 fused to eb1
25 d1l8ka_
not modelled
56.9
17
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Higher-molecular-weight phosphotyrosine protein phosphatases
26 c1l8kA_
not modelled
56.9
17
PDB header: hydrolaseChain: A: PDB Molecule: t-cell protein-tyrosine phosphatase;PDBTitle: t cell protein-tyrosine phosphatase structure
27 c4lxgA_
not modelled
56.4
19
PDB header: hydrolaseChain: A: PDB Molecule: mcp hydrolase;PDBTitle: crystal structure of dxnb2, a carbon - carbon bond hydrolase from2 sphingomonas wittichii rw1
28 c5n6yE_
not modelled
54.8
16
PDB header: oxidoreductaseChain: E: PDB Molecule: nitrogenase vanadium-iron protein beta chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
29 c2dt5A_
not modelled
54.1
16
PDB header: dna binding proteinChain: A: PDB Molecule: at-rich dna-binding protein;PDBTitle: crystal structure of ttha1657 (at-rich dna-binding protein) from2 thermus thermophilus hb8
30 d1bgva1
not modelled
52.7
32
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
31 c3fkkA_
not modelled
52.3
11
PDB header: lyaseChain: A: PDB Molecule: l-2-keto-3-deoxyarabonate dehydratase;PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
32 c1cr6A_
not modelled
52.1
24
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase;PDBTitle: crystal structure of murine soluble epoxide hydrolase2 complexed with cpu inhibitor
33 c3aogA_
not modelled
51.7
24
PDB header: oxidoreductaseChain: A: PDB Molecule: glutamate dehydrogenase;PDBTitle: crystal structure of glutamate dehydrogenase (gdhb) from thermus2 thermophilus (glu bound form)
34 c3r3jC_
not modelled
50.5
12
PDB header: oxidoreductaseChain: C: PDB Molecule: glutamate dehydrogenase;PDBTitle: kinetic and structural characterization of plasmodium falciparum2 glutamate dehydrogenase 2
35 c3s3fA_
not modelled
49.6
17
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: tyrosine-protein phosphatase 10d;PDBTitle: crystal structure of the catalytic domain of ptp10d from drosophila2 melanogaster with a small molecule inhibitor vanadate
36 c5sxyA_
not modelled
49.5
27
PDB header: chaperoneChain: A: PDB Molecule: bifunctional coenzyme pqq synthesis protein c/d;PDBTitle: the solution nmr structure for the pqqd truncation of methylobacterium2 extorquens pqqcd representing a functional and stand-alone3 ribosomally synthesized and post-translational modified (ripp)4 recognition element (rre)
37 c1v9lA_
not modelled
49.4
24
PDB header: oxidoreductaseChain: A: PDB Molecule: glutamate dehydrogenase;PDBTitle: l-glutamate dehydrogenase from pyrobaculum islandicum2 complexed with nad
38 c3i28A_
not modelled
49.4
24
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase 2;PDBTitle: crystal structure of soluble epoxide hydrolase
39 d1f74a_
not modelled
49.3
7
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
40 c5du2B_
not modelled
48.8
12
PDB header: transferaseChain: B: PDB Molecule: espg2 glycosyltransferase;PDBTitle: structural analysis of espg2 glycosyltransferase
41 c2p4sA_
not modelled
48.2
15
PDB header: transferaseChain: A: PDB Molecule: purine nucleoside phosphorylase;PDBTitle: structure of purine nucleoside phosphorylase from anopheles gambiae in2 complex with dadme-immh
42 c5xviA_
not modelled
47.8
25
PDB header: oxidoreductaseChain: A: PDB Molecule: glutamate dehydrogenase;PDBTitle: crystal structure of aspergillus niger apo- glutamate dehydrogenase
43 c3lerA_
not modelled
47.1
10
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from campylobacter2 jejuni subsp. jejuni nctc 11168
44 c3iaaB_
not modelled
47.1
11
PDB header: transferaseChain: B: PDB Molecule: calg2;PDBTitle: crystal structure of calg2, calicheamicin glycosyltransferase, tdp2 bound form
45 c3sboA_
not modelled
46.0
12
PDB header: oxidoreductaseChain: A: PDB Molecule: nadp-specific glutamate dehydrogenase;PDBTitle: structure of e.coli gdh from native source
46 d1o5ka_
not modelled
45.9
13
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
47 c1hrdA_
not modelled
45.9
32
PDB header: oxidoreductaseChain: A: PDB Molecule: glutamate dehydrogenase;PDBTitle: glutamate dehydrogenase
48 c5ijzH_
not modelled
45.5
15
PDB header: oxidoreductaseChain: H: PDB Molecule: nadp-specific glutamate dehydrogenase;PDBTitle: crystal strcuture of glutamate dehydrogenase(gdh) from corynebacterium2 glutamicum
49 c4n4qD_
not modelled
45.2
10
PDB header: lyaseChain: D: PDB Molecule: acylneuraminate lyase;PDBTitle: crystal structure of n-acetylneuraminate lyase from mycoplasma2 synoviae, crystal form ii
50 c1yn9B_
not modelled
44.6
19
PDB header: hydrolaseChain: B: PDB Molecule: polynucleotide 5'-phosphatase;PDBTitle: crystal structure of baculovirus rna 5'-phosphatase2 complexed with phosphate
51 c1u2eA_
not modelled
44.3
21
PDB header: hydrolaseChain: A: PDB Molecule: 2-hydroxy-6-ketonona-2,4-dienedioic acidPDBTitle: crystal structure of the c-c bond hydrolase mhpc
52 c4kyrA_
not modelled
44.2
33
PDB header: hydrolase, sugar binding proteinChain: A: PDB Molecule: phosphoglucan phosphatase lsf2, chloroplastic;PDBTitle: structure of a product bound plant phosphatase
53 c1nr1A_
not modelled
44.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: glutamate dehydrogenase 1;PDBTitle: crystal structure of the r463a mutant of human glutamate dehydrogenase
54 c3r3xA_
not modelled
43.5
22
PDB header: hydrolaseChain: A: PDB Molecule: fluoroacetate dehalogenase;PDBTitle: crystal structure of the fluoroacetate dehalogenase rpa1163 -2 asp110asn/bromoacetate
55 c2bzlA_
not modelled
43.2
20
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine-protein phosphatase, non-receptor type 14;PDBTitle: crystal structure of the human protein tyrosine phosphatase n14 at 1.2 65 a resolution
56 c4icnB_
not modelled
42.9
11
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: dihydrodipicolinate synthase from shewanella benthica
57 c4axvA_
not modelled
42.9
14
PDB header: hydrolaseChain: A: PDB Molecule: mpaa;PDBTitle: biochemical and structural characterization of the mpaa2 amidase as part of a conserved scavenging pathway for3 peptidoglycan derived peptides in gamma-proteobacteria
58 d1v9la1
not modelled
42.5
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
59 c4xgiA_
not modelled
42.5
24
PDB header: oxidoreductaseChain: A: PDB Molecule: glutamate dehydrogenase;PDBTitle: crystal structure of glutamate dehydrogenase from burkholderia2 thailandensis
60 c2c7sA_
not modelled
42.4
23
PDB header: hydrolaseChain: A: PDB Molecule: receptor-type tyrosine-protein phosphatase kappa;PDBTitle: crystal structure of human protein tyrosine phosphatase kappa at 1.95a2 resolution
61 c4d9jI_
not modelled
42.2
32
PDB header: de novo proteinChain: I: PDB Molecule: designed 16nm tetrahedral protein cage containing non-haemPDBTitle: structure of a 16 nm protein cage designed by fusing symmetric2 oligomeric domains
62 c4i19A_
not modelled
41.6
34
PDB header: hydrolaseChain: A: PDB Molecule: epoxide hydrolase;PDBTitle: the crystal structure of an epoxide hydrolase from streptomyces2 carzinostaticus subsp. neocarzinostaticus.
63 c3tqkA_
not modelled
41.5
13
PDB header: transferaseChain: A: PDB Molecule: phospho-2-dehydro-3-deoxyheptonate aldolase;PDBTitle: structure of phospho-2-dehydro-3-deoxyheptonate aldolase from2 francisella tularensis schu s4
64 c5kojD_
not modelled
41.4
15
PDB header: oxidoreductaseChain: D: PDB Molecule: nitrogenase femo beta subunit protein nifk;PDBTitle: nitrogenase mofep protein in the ids oxidized state
65 c3u7vA_
not modelled
41.2
26
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: the structure of a putative beta-galactosidase from caulobacter2 crescentus cb15.
66 c3k7dA_
not modelled
40.0
14
PDB header: transferaseChain: A: PDB Molecule: glutamate-ammonia-ligase adenylyltransferase;PDBTitle: c-terminal (adenylylation) domain of e.coli glutamine synthetase2 adenylyltransferase
67 d1larb1
not modelled
39.9
20
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Higher-molecular-weight phosphotyrosine protein phosphatases
68 d1ohea2
not modelled
39.9
23
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
69 c2bmaA_
not modelled
39.8
15
PDB header: oxidoreductaseChain: A: PDB Molecule: glutamate dehydrogenase (nadp+);PDBTitle: the crystal structure of plasmodium falciparum glutamate2 dehydrogenase, a putative target for novel antimalarial3 drugs
70 c3aoeC_
not modelled
39.2
21
PDB header: oxidoreductaseChain: C: PDB Molecule: glutamate dehydrogenase;PDBTitle: crystal structure of hetero-hexameric glutamate dehydrogenase from2 thermus thermophilus (leu bound form)
71 c3d0cB_
not modelled
39.2
11
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
72 c5bovD_
not modelled
38.8
18
PDB header: hydrolaseChain: D: PDB Molecule: putative epoxide hydrolase protein;PDBTitle: crystal structure of a putative epoxide hydrolase (kpn_01808) from2 klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 1.60 a3 resolution
73 d1rpma_
not modelled
38.7
26
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Higher-molecular-weight phosphotyrosine protein phosphatases
74 c2khzB_
not modelled
38.4
8
PDB header: nuclear proteinChain: B: PDB Molecule: c-myc-responsive protein rcl;PDBTitle: solution structure of rcl
75 c2jjdA_
not modelled
37.9
23
PDB header: hydrolaseChain: A: PDB Molecule: receptor-type tyrosine-protein phosphatase epsilon;PDBTitle: protein tyrosine phosphatase, receptor type, e isoform
76 c5xw4A_
not modelled
37.4
29
PDB header: cell cycleChain: A: PDB Molecule: tyrosine-protein phosphatase cdc14;PDBTitle: crystal structure of budding yeast cdc14p (wild type) in the apo state
77 c1tcvB_
not modelled
37.3
9
PDB header: transferaseChain: B: PDB Molecule: purine-nucleoside phosphorylase;PDBTitle: crystal structure of the purine nucleoside phosphorylase2 from schistosoma mansoni in complex with non-detergent3 sulfobetaine 195 and acetate
78 c1oheA_
not modelled
37.2
23
PDB header: hydrolaseChain: A: PDB Molecule: cdc14b2 phosphatase;PDBTitle: structure of cdc14b phosphatase with a peptide ligand
79 c4az1B_
not modelled
36.8
25
PDB header: hydrolaseChain: B: PDB Molecule: tyrosine specific protein phosphatase;PDBTitle: crystal structure of the trypanosoma cruzi protein tyrosine2 phosphatase tcptp1, a potential therapeutic target for chagas'3 disease
80 d1n8fa_
not modelled
36.6
17
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase
81 d1i9sa_
not modelled
36.6
27
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
82 d1lara1
not modelled
36.6
20
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Higher-molecular-weight phosphotyrosine protein phosphatases
83 c3n2xB_
not modelled
36.5
16
PDB header: lyaseChain: B: PDB Molecule: uncharacterized protein yage;PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
84 d1a3xa3
not modelled
36.5
13
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: Pyruvate kinase, C-terminal domain
85 d1h72c1
not modelled
35.9
12
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: GHMP Kinase, N-terminal domain
86 c3fobA_
not modelled
35.9
22
PDB header: oxidoreductaseChain: A: PDB Molecule: bromoperoxidase;PDBTitle: crystal structure of bromoperoxidase from bacillus anthracis
87 c5hxdB_
not modelled
35.7
16
PDB header: hydrolaseChain: B: PDB Molecule: protein mpaa;PDBTitle: crystal structure of murein-tripeptide amidase mpaa from escherichia2 coli o157
88 c3u9iA_
not modelled
35.6
8
PDB header: isomeraseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme, c-terminalPDBTitle: the crystal structure of mandelate racemase/muconate lactonizing2 enzyme from roseiflexus sp.
89 c2g59B_
not modelled
35.2
20
PDB header: hydrolaseChain: B: PDB Molecule: receptor-type tyrosine-protein phosphatase o;PDBTitle: crystal structure of the catalytic domain of protein2 tyrosine phosphatase from homo sapiens
90 d1s4ea1
not modelled
35.1
15
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: GHMP Kinase, N-terminal domain
91 c5f4zB_
not modelled
35.1
32
PDB header: hydrolaseChain: B: PDB Molecule: epoxide hydrolase;PDBTitle: the crystal structure of an epoxide hydrolase from streptomyces2 carzinostaticus subsp. neocarzinostaticus
92 c5egnB_
not modelled
34.8
13
PDB header: hydrolaseChain: B: PDB Molecule: esterase;PDBTitle: est816 as an n-acyl homoserine lactone degrading enzyme
93 c5ud6B_
not modelled
34.7
13
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dhdps from cyanidioschyzon merolae with lysine2 bound
94 c3sr9A_
not modelled
34.5
23
PDB header: hydrolaseChain: A: PDB Molecule: receptor-type tyrosine-protein phosphatase s;PDBTitle: crystal structure of mouse ptpsigma
95 c5jp6A_
not modelled
34.3
12
PDB header: hydrolaseChain: A: PDB Molecule: putative polysaccharide deacetylase;PDBTitle: bdellovibrio bacteriovorus peptidoglycan deacetylase bd3279
96 c1jpmB_
not modelled
34.0
17
PDB header: isomeraseChain: B: PDB Molecule: l-ala-d/l-glu epimerase;PDBTitle: l-ala-d/l-glu epimerase
97 c4ah7C_
not modelled
34.0
7
PDB header: lyaseChain: C: PDB Molecule: n-acetylneuraminate lyase;PDBTitle: structure of wild type stapylococcus aureus n-acetylneuraminic acid2 lyase in complex with pyruvate
98 c3wg9D_
not modelled
33.6
18
PDB header: transcriptionChain: D: PDB Molecule: redox-sensing transcriptional repressor rex;PDBTitle: crystal structure of rsp, a rex-family repressor
99 c2c46B_
not modelled
33.1
27
PDB header: transferaseChain: B: PDB Molecule: mrna capping enzyme;PDBTitle: crystal structure of the human rna guanylyltransferase and 5'-2 phosphatase
100 c2nz6A_
not modelled
33.0
31
PDB header: hydrolaseChain: A: PDB Molecule: receptor-type tyrosine-protein phosphatase eta;PDBTitle: crystal structure of the ptprj inactivating mutant c1239s
101 d1wota_
not modelled
32.9
23
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase
102 d1yfoa_
not modelled
32.4
17
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Higher-molecular-weight phosphotyrosine protein phosphatases
103 c3dz1A_
not modelled
32.2
12
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
104 c3e96B_
not modelled
32.0
9
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from bacillus2 clausii
105 c5b3fB_
not modelled
31.6
22
PDB header: transferaseChain: B: PDB Molecule: phosphoribulokinase/uridine kinase;PDBTitle: crystal structure of phosphoribulokinase from methanospirillum2 hungatei
106 c3k8zD_
not modelled
31.1
21
PDB header: oxidoreductaseChain: D: PDB Molecule: nad-specific glutamate dehydrogenase;PDBTitle: crystal structure of gudb1 a decryptified secondary glutamate2 dehydrogenase from b. subtilis
107 c4nyhB_
not modelled
30.4
17
PDB header: hydrolaseChain: B: PDB Molecule: rna/rnp complex-1-interacting phosphatase;PDBTitle: orthorhombic crystal form of pir1 dual specificity phosphatase core
108 c2rfgB_
not modelled
30.3
9
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
109 d1p15a_
not modelled
30.2
27
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Higher-molecular-weight phosphotyrosine protein phosphatases
110 d1xxxa1
not modelled
30.2
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
111 c3tjiA_
not modelled
30.1
14
PDB header: lyaseChain: A: PDB Molecule: mandelate racemase/muconate lactonizing enzyme, n-terminalPDBTitle: crystal structure of an enolase from enterobacter sp. 638 (efi target2 efi-501662) with bound mg
112 c4ur7B_
not modelled
30.1
5
PDB header: lyaseChain: B: PDB Molecule: keto-deoxy-d-galactarate dehydratase;PDBTitle: crystal structure of keto-deoxy-d-galactarate dehydratase2 complexed with pyruvate
113 c6ckgA_
not modelled
29.8
13
PDB header: transferaseChain: A: PDB Molecule: d-glycerate 3-kinase;PDBTitle: d-glycerate 3-kinase from cryptococcus neoformans var. grubii serotype2 a (h99 / atcc 208821 / cbs 10515 / fgsc 9487)
114 d1hkha_
not modelled
29.7
25
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Haloperoxidase
115 c2m3vA_
not modelled
29.5
25
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of tyrosine phosphatase related to biofilm2 formation a (tpba) from pseudomonas aeruginosa
116 c3othB_
not modelled
29.4
12
PDB header: transferase/antibioticChain: B: PDB Molecule: calg1;PDBTitle: crystal structure of calg1, calicheamicin glycostyltransferase, tdp2 and calicheamicin alpha3i bound form
117 c5cxsA_
not modelled
29.2
13
PDB header: transferaseChain: A: PDB Molecule: purine nucleoside phosphorylase;PDBTitle: crystal structure of isoform 2 of purine nucleoside phosphorylase2 complexed with mes
118 c4xkyC_
not modelled
29.2
17
PDB header: lyaseChain: C: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: structure of dihydrodipicolinate synthase from the commensal bacterium2 bacteroides thetaiotaomicron at 2.1 a resolution
119 c3wibB_
not modelled
29.2
15
PDB header: hydrolaseChain: B: PDB Molecule: haloalkane dehalogenase;PDBTitle: crystal structure of y109w mutant haloalkane dehalogenase data from2 agrobacterium tumefaciens c58
120 d1mkpa_
not modelled
29.2
29
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like