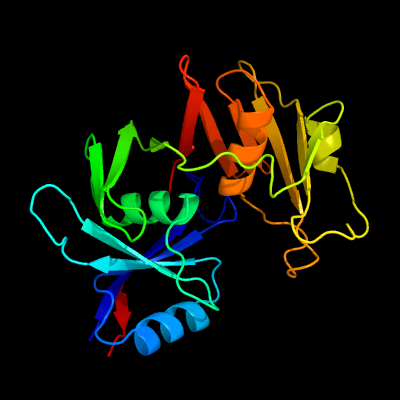
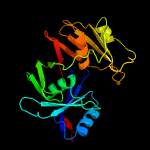
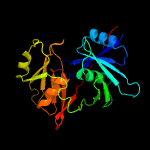
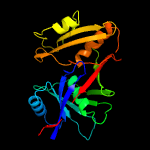
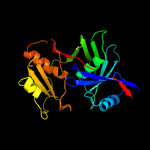
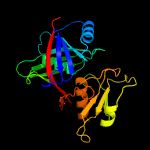
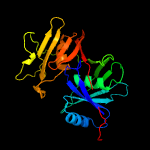
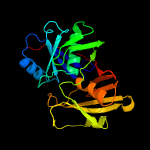
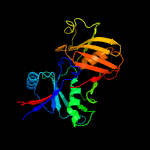
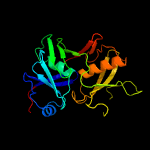
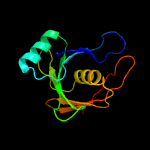
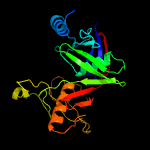
1 c1ym5A_
100.0
31
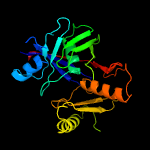
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical 32.6 kda protein in dap2-slt2PDBTitle: crystal structure of yhi9, the yeast member of the2 phenazine biosynthesis phzf enzyme superfamily.
2 c3ednB_
100.0
26
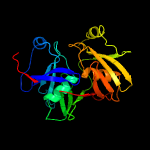
PDB header: biosynthetic proteinChain: B: PDB Molecule: phenazine biosynthesis protein, phzf family;PDBTitle: crystal structure of the bacillus anthracis phenazine2 biosynthesis protein, phzf family
3 c4dunA_
100.0
23
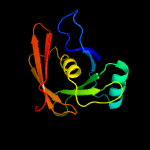
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative phenazine biosynthesis phzc/phzf protein;PDBTitle: 1.76a x-ray crystal structure of a putative phenazine biosynthesis2 phzc/phzf protein from clostridium difficile (strain 630)
4 d1s7ja_
100.0
20
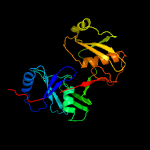
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
5 c1u0kA_
100.0
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: gene product pa4716;PDBTitle: the structure of a predicted epimerase pa4716 from pseudomonas2 aeruginosa
6 c1qy9B_
100.0
24
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein ydde;PDBTitle: crystal structure of e. coli se-met protein ydde
7 c1u1wA_
100.0
31
PDB header: isomerase, lyaseChain: A: PDB Molecule: phenazine biosynthesis protein phzf;PDBTitle: structure and function of phenazine-biosynthesis protein phzf from2 pseudomonas fluorescens 2-79
8 c5ha4A_
100.0
23
PDB header: isomeraseChain: A: PDB Molecule: diaminopimelate epimerase;PDBTitle: structure of a diaminopimelate epimerase from acinetobacter baumannii
9 c3ekmE_
100.0
18
PDB header: isomeraseChain: E: PDB Molecule: diaminopimelate epimerase, chloroplastic;PDBTitle: crystal structure of diaminopimelate epimerase form2 arabidopsis thaliana in complex with irreversible inhibitor3 dl-azidap
10 d1u0ka1
100.0
26
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
11 c4juuA_
100.0
22
PDB header: isomeraseChain: A: PDB Molecule: epimerase;PDBTitle: crystal structure of a putative hydroxyproline epimerase from2 xanthomonas campestris (target efi-506516) with bound phosphate and3 unknown ligand
12 c6r77B_
100.0
18
PDB header: lyaseChain: B: PDB Molecule: proline racemase;PDBTitle: crystal structure of trans-3-hydroxy-l-proline dehydratase in complex2 with substrate - closed conformation
13 d1xuba1
100.0
26
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
14 c6j7cA_
100.0
16
PDB header: isomeraseChain: A: PDB Molecule: proline racemase;PDBTitle: crystal structure of proline racemase-like protein from thermococcus2 litoralis in complex with proline
15 c4jd7D_
100.0
19
PDB header: isomeraseChain: D: PDB Molecule: proline racemase;PDBTitle: crystal structure of pput_1285, a putative hydroxyproline epimerase2 from pseudomonas putida f1 (target efi-506500), open form, space3 group p212121, bound sulfate
16 c2azpA_
100.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pa1268;PDBTitle: crystal structure of pa1268 solved by sulfur sad
17 c2gkjA_
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: diaminopimelate epimerase;PDBTitle: crystal structure of diaminopimelate epimerase in complex2 with an irreversible inhibitor dl-azidap
18 d1qy9a1
100.0
25
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
19 c2otnB_
100.0
20
PDB header: isomeraseChain: B: PDB Molecule: diaminopimelate epimerase;PDBTitle: crystal structure of the catalytically active form of diaminopimelate2 epimerase from bacillus anthracis
20 c4k7gD_
100.0
21
PDB header: isomeraseChain: D: PDB Molecule: 3-hydroxyproline dehydratse;PDBTitle: crystal structure of a 3-hydroxyproline dehydratse from agrobacterium2 vitis, target efi-506470, with bound pyrrole 2-carboxylate, ordered3 active site
21 c1w62B_
not modelled
100.0
18
PDB header: racemaseChain: B: PDB Molecule: proline racemase a;PDBTitle: proline racemase in complex with one molecule of pyrrole-2-carboxylic2 acid (hemi form)
22 c6hjgB_
not modelled
100.0
17
PDB header: isomeraseChain: B: PDB Molecule: proline racemase a;PDBTitle: trypanosoma cruzi proline racemase in complex with inhibitor oxopa
23 c4lb0B_
not modelled
99.9
19
PDB header: isomeraseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a hydroxyproline epimerase from agrobacterium2 vitis, target efi-506420, with bound trans-4-oh-l-proline
24 d1qy9a2
not modelled
99.9
20
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
25 d1xuba2
not modelled
99.9
33
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
26 d1tm0a_
not modelled
99.8
17
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: Proline racemase
27 d1u0ka2
not modelled
99.8
27
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
28 d2gkea1
not modelled
99.6
14
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: Diaminopimelate epimerase
29 c5m47A_
not modelled
99.4
15
PDB header: isomeraseChain: A: PDB Molecule: diaminopimelate epimerase;PDBTitle: crystal strcuture of dapf from corynebacterium glutamicum in complex2 with d,l-diaminopimelate
30 c3fveA_
not modelled
99.3
18
PDB header: isomeraseChain: A: PDB Molecule: diaminopimelate epimerase;PDBTitle: crystal structure of diaminopimelate epimerase mycobacterium2 tuberculosis dapf
31 d2gkea2
not modelled
97.2
17
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: Diaminopimelate epimerase
32 c6p3hB_
not modelled
90.7
18
PDB header: isomeraseChain: B: PDB Molecule: (4e)-oxalomesaconate delta-isomerase;PDBTitle: crystal structure of ligu(k66m) bound to substrate
33 c6otvA_
not modelled
86.5
18
PDB header: isomeraseChain: A: PDB Molecule: putative isomerase ybhh;PDBTitle: crystal structure of putative isomerase ec2056
34 d2h9fa2
not modelled
82.7
21
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PA0793-like
35 c2pw0A_
not modelled
71.5
22
PDB header: unknown functionChain: A: PDB Molecule: prpf methylaconitate isomerase;PDBTitle: crystal structure of trans-aconitate bound to methylaconitate2 isomerase prpf from shewanella oneidensis
36 c3g7kD_
not modelled
62.1
27
PDB header: isomeraseChain: D: PDB Molecule: 3-methylitaconate isomerase;PDBTitle: crystal structure of methylitaconate-delta-isomerase
37 c5hr4J_
not modelled
45.6
29
PDB header: hydrolase/dnaChain: J: PDB Molecule: mmei;PDBTitle: structure of type iil restriction-modification enzyme mmei in complex2 with dna has implications for engineering of new specificities
38 c5z2tC_
not modelled
25.3
29
PDB header: dna binding protein/dnaChain: C: PDB Molecule: double homeobox protein 4;PDBTitle: crystal structure of dna-bound dux4-hd2
39 d1uhsa_
not modelled
22.8
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
40 d1up8a_
not modelled
21.9
16
Fold: Acid phosphatase/Vanadium-dependent haloperoxidaseSuperfamily: Acid phosphatase/Vanadium-dependent haloperoxidaseFamily: Haloperoxidase (bromoperoxidase)
41 d2cuea1
not modelled
18.8
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
42 d1yh5a1
not modelled
18.8
9
Fold: YggU-likeSuperfamily: YggU-likeFamily: YggU-like
43 c1x2mA_
not modelled
18.5
12
PDB header: transcriptionChain: A: PDB Molecule: lag1 longevity assurance homolog 6;PDBTitle: solution structure of the homeobox domain of mouse lag12 longevity assurance homolog 6
44 d1s7ea1
not modelled
17.8
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
45 c2da2A_
not modelled
17.7
12
PDB header: transcriptionChain: A: PDB Molecule: alpha-fetoprotein enhancer binding protein;PDBTitle: solution structure of the second homeobox domain of at-2 binding transcription factor 1 (atbf1)
46 c2waqG_
not modelled
17.3
31
PDB header: transcriptionChain: G: PDB Molecule: dna-directed rna polymerase rpo8 subunit;PDBTitle: the complete structure of the archaeal 13-subunit dna-directed rna2 polymerase
47 c2dmuA_
not modelled
17.1
29
PDB header: dna binding proteinChain: A: PDB Molecule: homeobox protein goosecoid;PDBTitle: solution structure of the homeobox domain of homeobox2 protein goosecoid
48 d2p81a1
not modelled
17.0
41
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
49 c6aziA_
not modelled
16.4
19
PDB header: hydrolaseChain: A: PDB Molecule: d-alanyl-d-alanine endopeptidase;PDBTitle: 1.75 angstrom resolution crystal structure of d-alanyl-d-alanine2 endopeptidase from enterobacter cloacae in complex with covalently3 bound boronic acid
50 d1ftza_
not modelled
16.3
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
51 c2l9rA_
not modelled
16.0
28
PDB header: transcriptionChain: A: PDB Molecule: homeobox protein nkx-3.1;PDBTitle: solution nmr structure of homeobox domain of homeobox protein nkx-3.12 from homo sapiens, northeast structural genomics consortium target3 hr6470a
52 d1o4xa1
not modelled
15.9
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
53 d1e3oc1
not modelled
15.7
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
54 d2ar0a1
not modelled
15.6
36
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N-6 DNA Methylase-like
55 d1au7a1
not modelled
15.4
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
56 d1o9ga_
not modelled
15.4
16
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: rRNA methyltransferase AviRa
57 d2craa1
not modelled
14.7
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
58 c3k0bA_
not modelled
14.5
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted n6-adenine-specific dna methylase;PDBTitle: crystal structure of a predicted n6-adenine-specific dna methylase2 from listeria monocytogenes str. 4b f2365
59 d1p7ia_
not modelled
14.2
41
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
60 d1le8a_
not modelled
14.0
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
61 c3ufbA_
not modelled
14.0
21
PDB header: transferaseChain: A: PDB Molecule: type i restriction-modification system methyltransferasePDBTitle: crystal structure of a modification subunit of a putative type i2 restriction enzyme from vibrio vulnificus yj016
62 c2da4A_
not modelled
13.9
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein dkfzp686k21156;PDBTitle: solution structure of the homeobox domain of the2 hypothetical protein, dkfzp686k21156
63 c2vi6F_
not modelled
13.8
18
PDB header: transcriptionChain: F: PDB Molecule: homeobox protein nanog;PDBTitle: crystal structure of the nanog homeodomain
64 c3khkA_
not modelled
13.6
21
PDB header: dna binding proteinChain: A: PDB Molecule: type i restriction-modification system methylation subunit;PDBTitle: crystal structure of type-i restriction-modification system2 methylation subunit (mm_0429) from methanosarchina mazei.
65 c3a01A_
not modelled
13.5
17
PDB header: gene regulation/dnaChain: A: PDB Molecule: homeodomain-containing protein;PDBTitle: crystal structure of aristaless and clawless homeodomains bound to dna
66 d1ig7a_
not modelled
13.4
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
67 d2hi3a1
not modelled
13.3
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
68 c5hfjF_
not modelled
13.3
14
PDB header: dna binding proteinChain: F: PDB Molecule: adenine specific dna methyltransferase (dpna);PDBTitle: crystal structure of m1.hpyavi-sam complex
69 c2da1A_
not modelled
13.2
18
PDB header: transcriptionChain: A: PDB Molecule: alpha-fetoprotein enhancer binding protein;PDBTitle: solution structure of the first homeobox domain of at-2 binding transcription factor 1 (atbf1)
70 d1octc1
not modelled
13.2
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
71 d2cqxa1
not modelled
12.8
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
72 d1jgga_
not modelled
12.8
41
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
73 d1fjlb_
not modelled
12.5
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
74 d2okca1
not modelled
12.1
36
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N-6 DNA Methylase-like
75 c2dmsA_
not modelled
12.0
17
PDB header: dna binding proteinChain: A: PDB Molecule: homeobox protein otx2;PDBTitle: solution structure of the homeobox domain of homeobox2 protein otx2
76 c2e19A_
not modelled
12.0
6
PDB header: transcriptionChain: A: PDB Molecule: transcription factor 8;PDBTitle: solution structure of the homeobox domain from human nil-2-2 a zinc finger protein, transcription factor 8
77 d1g60a_
not modelled
11.9
14
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Type II DNA methylase
78 d1f43a_
not modelled
11.9
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
79 c5ybbA_
not modelled
11.8
29
PDB header: dna binding protein/dnaChain: A: PDB Molecule: type i restriction-modification system methyltransferasePDBTitle: structural basis underlying complex assembly andconformational2 transition of the type i r-m system
80 c4h4nA_
not modelled
11.8
29
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein ba_2335;PDBTitle: 1.1 angstrom crystal structure of hypothetical protein ba_2335 from2 bacillus anthracis
81 d1zq3p1
not modelled
11.7
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
82 d2r5yb1
not modelled
11.6
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
83 c3itcA_
not modelled
11.4
14
PDB header: hydrolaseChain: A: PDB Molecule: renal dipeptidase;PDBTitle: crystal structure of sco3058 with bound citrate and glycerol
84 c2zifB_
not modelled
11.3
20
PDB header: transferaseChain: B: PDB Molecule: putative modification methylase;PDBTitle: crystal structure of ttha0409, putative dna modification methylase2 from thermus thermophilus hb8- complexed with s-adenosyl-l-methionine
85 d1bgva2
not modelled
11.0
13
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases
86 d2hddb_
not modelled
10.9
41
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
87 d1b8ia_
not modelled
10.8
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
88 d9anta_
not modelled
10.6
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
89 c2k40A_
not modelled
10.6
12
PDB header: dna binding proteinChain: A: PDB Molecule: homeobox expressed in es cells 1;PDBTitle: nmr structure of hesx-1 homeodomain double mutant r31l/e42l
90 c1g38A_
not modelled
10.5
7
PDB header: transferase/dnaChain: A: PDB Molecule: modification methylase taqi;PDBTitle: adenine-specific methyltransferase m. taq i/dna complex
91 d1vnda_
not modelled
10.4
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
92 c2i5gB_
not modelled
10.4
14
PDB header: hydrolaseChain: B: PDB Molecule: amidohydrolase;PDBTitle: crystal strcuture of amidohydrolase from pseudomonas aeruginosa
93 c5cffE_
not modelled
10.4
13
PDB header: transcription/rna binding proteinChain: E: PDB Molecule: staufen;PDBTitle: crystal structure of miranda/staufen dsrbd5 complex
94 c3a03A_
not modelled
10.3
18
PDB header: gene regulationChain: A: PDB Molecule: t-cell leukemia homeobox protein 2;PDBTitle: crystal structure of hox11l1 homeodomain
95 c3ldgA_
not modelled
10.1
14
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein smu.472;PDBTitle: crystal structure of smu.472, a putative methyltransferase complexed2 with sah
96 c1aqjB_
not modelled
10.1
7
PDB header: methyltransferaseChain: B: PDB Molecule: adenine-n6-dna-methyltransferase taqi;PDBTitle: structure of adenine-n6-dna-methyltransferase taqi
97 d1du0a_
not modelled
10.0
41
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain
98 c3lu2B_
not modelled
10.0
0
PDB header: hydrolaseChain: B: PDB Molecule: lmo2462 protein;PDBTitle: structure of lmo2462, a listeria monocytogenes amidohydrolase family2 putative dipeptidase
99 c2ragB_
not modelled
9.9
14
PDB header: hydrolaseChain: B: PDB Molecule: dipeptidase;PDBTitle: crystal structure of aminohydrolase from caulobacter crescentus