| 1 |
|
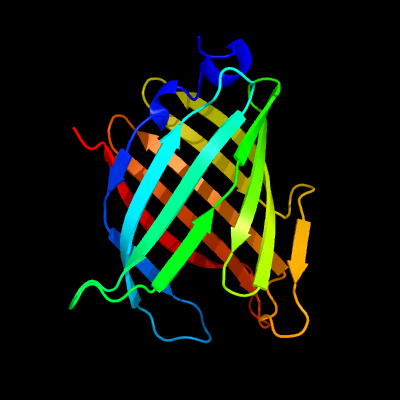
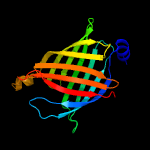
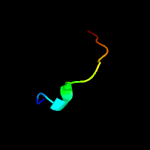
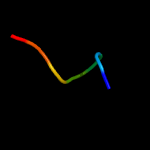
PDB 2fr2 chain A domain 1
Region: 4 - 164
Aligned: 161
Modelled: 161
Confidence: 100.0%
Identity: 100%
Fold: Lipocalins
Superfamily: Lipocalins
Family: Rv2717c-like
Phyre2
| 2 |
|
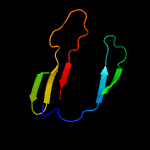
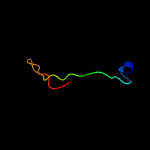
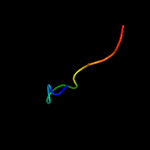
PDB 2a13 chain A domain 1
Region: 3 - 163
Aligned: 153
Modelled: 161
Confidence: 100.0%
Identity: 41%
Fold: Lipocalins
Superfamily: Lipocalins
Family: Rv2717c-like
Phyre2
| 3 |
|
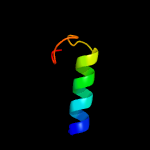
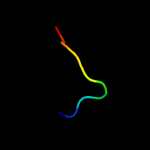
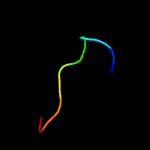
PDB 2fwv chain A
Region: 3 - 164
Aligned: 152
Modelled: 162
Confidence: 100.0%
Identity: 32%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein mtubf_01000852;
PDBTitle: crystal structure of rv0813
Phyre2
| 4 |
|
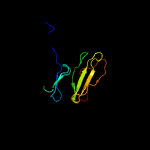
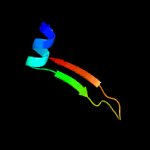
PDB 3ia8 chain A
Region: 3 - 163
Aligned: 156
Modelled: 161
Confidence: 100.0%
Identity: 32%
PDB header:metal binding protein
Chain: A: PDB Molecule:thap domain-containing protein 4;
PDBTitle: the structure of the c-terminal heme nitrobindin domain of thap2 domain-containing protein 4 from homo sapiens
Phyre2
| 5 |
|
PDB 2o62 chain A domain 1
Region: 12 - 71
Aligned: 56
Modelled: 60
Confidence: 73.5%
Identity: 32%
Fold: Lipocalins
Superfamily: Lipocalins
Family: All1756-like
Phyre2
| 6 |
|
PDB 1jva chain A domain 2
Region: 7 - 30
Aligned: 24
Modelled: 24
Confidence: 45.7%
Identity: 33%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Intein endonuclease
Phyre2
| 7 |
|
PDB 5yzn chain A
Region: 28 - 125
Aligned: 97
Modelled: 98
Confidence: 37.9%
Identity: 11%
PDB header:hydrolase
Chain: A: PDB Molecule:acyl-peptide hydrolase, putative;
PDBTitle: crystal structure of s9 peptidase (active form) from deinococcus2 radiodurans r1
Phyre2
| 8 |
|
PDB 4mve chain B
Region: 10 - 23
Aligned: 14
Modelled: 14
Confidence: 27.4%
Identity: 29%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of tcur_1030 protein from thermomonospora curvata
Phyre2
| 9 |
|
PDB 5yvf chain D
Region: 12 - 37
Aligned: 26
Modelled: 26
Confidence: 17.6%
Identity: 31%
PDB header:plant protein
Chain: D: PDB Molecule:bfa1;
PDBTitle: crystal structure of bfa1
Phyre2
| 10 |
|
PDB 2d8z chain A domain 1
Region: 61 - 68
Aligned: 8
Modelled: 8
Confidence: 8.7%
Identity: 38%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 11 |
|
PDB 4kkp chain A
Region: 52 - 71
Aligned: 20
Modelled: 20
Confidence: 8.1%
Identity: 30%
PDB header:structural protein
Chain: A: PDB Molecule:rbma protein;
PDBTitle: crystal structure of vibrio cholerae rbma (crystal form 2)
Phyre2
| 12 |
|
PDB 3fqm chain A
Region: 17 - 26
Aligned: 10
Modelled: 10
Confidence: 7.1%
Identity: 40%
PDB header:metal binding protein
Chain: A: PDB Molecule:non-structural protein 5a;
PDBTitle: crystal structure of a novel dimeric form of hcv ns5a domain i protein
Phyre2
| 13 |
|
PDB 1b24 chain A
Region: 13 - 42
Aligned: 30
Modelled: 30
Confidence: 6.8%
Identity: 23%
PDB header:intron-encoded
Chain: A: PDB Molecule:protein (i-dmoi);
PDBTitle: i-dmoi, intron-encoded endonuclease
Phyre2
| 14 |
|
PDB 6dzp chain G
Region: 1 - 15
Aligned: 15
Modelled: 14
Confidence: 6.4%
Identity: 13%
PDB header:ribosome
Chain: G: PDB Molecule:50s ribosomal protein l6;
PDBTitle: cryo-em structure of mycobacterium smegmatis c(minus) 50s ribosomal2 subunit
Phyre2
| 15 |
|
PDB 2cur chain A domain 2
Region: 61 - 68
Aligned: 8
Modelled: 8
Confidence: 6.2%
Identity: 25%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 16 |
|
PDB 4i0x chain A
Region: 15 - 21
Aligned: 7
Modelled: 7
Confidence: 6.1%
Identity: 57%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:esat-6-like protein mab_3112;
PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
Phyre2
| 17 |
|
PDB 1vs6 chain Z domain 1
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 5.6%
Identity: 23%
Fold: L28p-like
Superfamily: L28p-like
Family: Ribosomal protein L31p
Phyre2
| 18 |
|
PDB 2b66 chain 4
Region: 1 - 15
Aligned: 15
Modelled: 15
Confidence: 5.6%
Identity: 27%
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: 50s ribosomal subunit from a crystal structure of release factor rf1,2 trnas and mrna bound to the ribosome. this file contains the 50s3 subunit from a crystal structure of release factor rf1, trnas and4 mrna bound to the ribosome and is described in remark 400
Phyre2
| 19 |
|
PDB 2j03 chain 4
Region: 1 - 12
Aligned: 12
Modelled: 12
Confidence: 5.3%
Identity: 25%
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 4 of 4).3 this file contains the 50s subunit from molecule ii.
Phyre2
| 20 |
|
PDB 2j01 chain 4 domain 1
Region: 1 - 12
Aligned: 12
Modelled: 12
Confidence: 5.3%
Identity: 25%
Fold: L28p-like
Superfamily: L28p-like
Family: Ribosomal protein L31p
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|