| 1 | c3fveA_
|
|
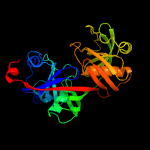 |
100.0 |
96 |
PDB header:isomerase
Chain: A: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of diaminopimelate epimerase mycobacterium2 tuberculosis dapf
|
|
|
|
| 2 | c5m47A_
|
|
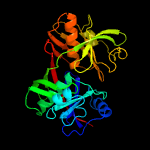 |
100.0 |
53 |
PDB header:isomerase
Chain: A: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal strcuture of dapf from corynebacterium glutamicum in complex2 with d,l-diaminopimelate
|
|
|
|
| 3 | c2gkjA_
|
|
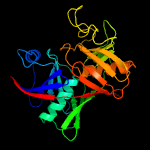 |
100.0 |
28 |
PDB header:isomerase
Chain: A: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of diaminopimelate epimerase in complex2 with an irreversible inhibitor dl-azidap
|
|
|
|
| 4 | c5ha4A_
|
|
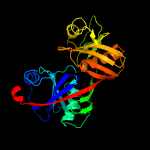 |
100.0 |
31 |
PDB header:isomerase
Chain: A: PDB Molecule:diaminopimelate epimerase;
PDBTitle: structure of a diaminopimelate epimerase from acinetobacter baumannii
|
|
|
|
| 5 | c2otnB_
|
|
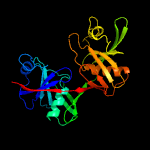 |
100.0 |
31 |
PDB header:isomerase
Chain: B: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of the catalytically active form of diaminopimelate2 epimerase from bacillus anthracis
|
|
|
|
| 6 | c3ekmE_
|
|
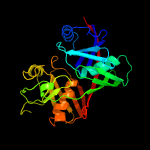 |
100.0 |
35 |
PDB header:isomerase
Chain: E: PDB Molecule:diaminopimelate epimerase, chloroplastic;
PDBTitle: crystal structure of diaminopimelate epimerase form2 arabidopsis thaliana in complex with irreversible inhibitor3 dl-azidap
|
|
|
|
| 7 | d2gkea1
|
|
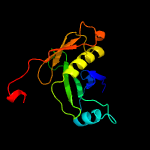 |
100.0 |
32 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:Diaminopimelate epimerase |
|
|
|
| 8 | c4juuA_
|
|
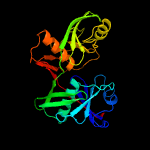 |
100.0 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:epimerase;
PDBTitle: crystal structure of a putative hydroxyproline epimerase from2 xanthomonas campestris (target efi-506516) with bound phosphate and3 unknown ligand
|
|
|
|
| 9 | c3ednB_
|
|
 |
100.0 |
17 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:phenazine biosynthesis protein, phzf family;
PDBTitle: crystal structure of the bacillus anthracis phenazine2 biosynthesis protein, phzf family
|
|
|
|
| 10 | c1qy9B_
|
|
 |
100.0 |
17 |
PDB header:unknown function
Chain: B: PDB Molecule:hypothetical protein ydde;
PDBTitle: crystal structure of e. coli se-met protein ydde
|
|
|
|
| 11 | d1s7ja_
|
|
 |
100.0 |
15 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
|
|
|
| 12 | c1ym5A_
|
|
 |
100.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical 32.6 kda protein in dap2-slt2
PDBTitle: crystal structure of yhi9, the yeast member of the2 phenazine biosynthesis phzf enzyme superfamily.
|
|
|
|
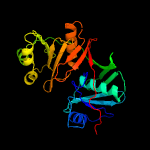
| 13 | c4dunA_
|
|
 |
100.0 |
14 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative phenazine biosynthesis phzc/phzf protein;
PDBTitle: 1.76a x-ray crystal structure of a putative phenazine biosynthesis2 phzc/phzf protein from clostridium difficile (strain 630)
|
|
|
|
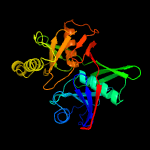
| 14 | c2azpA_
|
|
 |
100.0 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pa1268;
PDBTitle: crystal structure of pa1268 solved by sulfur sad
|
|
|
|
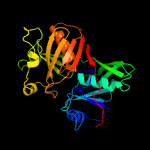
| 15 | c1u1wA_
|
|
 |
100.0 |
15 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:phenazine biosynthesis protein phzf;
PDBTitle: structure and function of phenazine-biosynthesis protein phzf from2 pseudomonas fluorescens 2-79
|
|
|
|
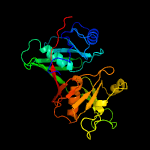
| 16 | c1u0kA_
|
|
 |
100.0 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:gene product pa4716;
PDBTitle: the structure of a predicted epimerase pa4716 from pseudomonas2 aeruginosa
|
|
|
|
| 17 | c4jd7D_
|
|
 |
100.0 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:proline racemase;
PDBTitle: crystal structure of pput_1285, a putative hydroxyproline epimerase2 from pseudomonas putida f1 (target efi-506500), open form, space3 group p212121, bound sulfate
|
|
|
|
| 18 | c6r77B_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:proline racemase;
PDBTitle: crystal structure of trans-3-hydroxy-l-proline dehydratase in complex2 with substrate - closed conformation
|
|
|
|
| 19 | d1tm0a_
|
|
 |
100.0 |
13 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:Proline racemase |
|
|
|
| 20 | c6hjgB_
|
|
 |
100.0 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:proline racemase a;
PDBTitle: trypanosoma cruzi proline racemase in complex with inhibitor oxopa
|
|
|
|
| 21 | c1w62B_ |
|
not modelled |
100.0 |
16 |
PDB header:racemase
Chain: B: PDB Molecule:proline racemase a;
PDBTitle: proline racemase in complex with one molecule of pyrrole-2-carboxylic2 acid (hemi form)
|
|
|
| 22 | c6j7cA_ |
|
not modelled |
100.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:proline racemase;
PDBTitle: crystal structure of proline racemase-like protein from thermococcus2 litoralis in complex with proline
|
|
|
| 23 | c4lb0B_ |
|
not modelled |
100.0 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a hydroxyproline epimerase from agrobacterium2 vitis, target efi-506420, with bound trans-4-oh-l-proline
|
|
|
| 24 | d2gkea2 |
|
not modelled |
100.0 |
23 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:Diaminopimelate epimerase |
|
|
| 25 | c4k7gD_ |
|
not modelled |
100.0 |
15 |
PDB header:isomerase
Chain: D: PDB Molecule:3-hydroxyproline dehydratse;
PDBTitle: crystal structure of a 3-hydroxyproline dehydratse from agrobacterium2 vitis, target efi-506470, with bound pyrrole 2-carboxylate, ordered3 active site
|
|
|
| 26 | d1qy9a1 |
|
not modelled |
99.8 |
18 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
|
|
| 27 | d1xuba1 |
|
not modelled |
99.7 |
12 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
|
|
| 28 | d1xuba2 |
|
not modelled |
99.6 |
17 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
|
|
| 29 | d1qy9a2 |
|
not modelled |
99.6 |
16 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
|
|
| 30 | d1u0ka2 |
|
not modelled |
99.5 |
17 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
|
|
| 31 | d1u0ka1 |
|
not modelled |
99.4 |
13 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PhzC/PhzF-like |
|
|
| 32 | c6p3hB_ |
|
not modelled |
98.4 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:(4e)-oxalomesaconate delta-isomerase;
PDBTitle: crystal structure of ligu(k66m) bound to substrate
|
|
|
| 33 | c6otvA_ |
|
not modelled |
98.3 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:putative isomerase ybhh;
PDBTitle: crystal structure of putative isomerase ec2056
|
|
|
| 34 | c3g7kD_ |
|
not modelled |
97.9 |
15 |
PDB header:isomerase
Chain: D: PDB Molecule:3-methylitaconate isomerase;
PDBTitle: crystal structure of methylitaconate-delta-isomerase
|
|
|
| 35 | c2pw0A_ |
|
not modelled |
97.1 |
19 |
PDB header:unknown function
Chain: A: PDB Molecule:prpf methylaconitate isomerase;
PDBTitle: crystal structure of trans-aconitate bound to methylaconitate2 isomerase prpf from shewanella oneidensis
|
|
|
| 36 | d1sr8a_ |
|
not modelled |
56.4 |
39 |
Fold:CbiD-like
Superfamily:CbiD-like
Family:CbiD-like |
|
|
| 37 | d2h9fa2 |
|
not modelled |
55.6 |
23 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PA0793-like |
|
|
| 38 | c3axtA_ |
|
not modelled |
31.9 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:probable n(2),n(2)-dimethylguanosine trna methyltransferase
PDBTitle: complex structure of trna methyltransferase trm1 from aquifex aeolicus2 with s-adenosyl-l-methionine
|
|
|
| 39 | d2h9fa1 |
|
not modelled |
25.8 |
13 |
Fold:Diaminopimelate epimerase-like
Superfamily:Diaminopimelate epimerase-like
Family:PA0793-like |
|
|
| 40 | d2dula1 |
|
not modelled |
23.2 |
33 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TRM1-like |
|
|
| 41 | c5jvoA_ |
|
not modelled |
20.9 |
21 |
PDB header:dna binding protein
Chain: A: PDB Molecule:arginine repressor;
PDBTitle: crystal structure of the arginine repressor from the pathogenic2 bacterium corynebacterium pseudotuberculosis
|
|
|
| 42 | c5hr4J_ |
|
not modelled |
20.6 |
17 |
PDB header:hydrolase/dna
Chain: J: PDB Molecule:mmei;
PDBTitle: structure of type iil restriction-modification enzyme mmei in complex2 with dna has implications for engineering of new specificities
|
|
|
| 43 | d1vema1 |
|
not modelled |
19.5 |
11 |
Fold:Prealbumin-like
Superfamily:Starch-binding domain-like
Family:Starch-binding domain |
|
|
| 44 | d2ih2a1 |
|
not modelled |
19.2 |
18 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:DNA methylase TaqI, N-terminal domain |
|
|
| 45 | c4o1hA_ |
|
not modelled |
19.0 |
27 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcription regulator glnr;
PDBTitle: crystal structure of the regulatory domain of ameglnr
|
|
|
| 46 | d1q7sa_ |
|
not modelled |
18.7 |
16 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
|
|
| 47 | d2ar0a1 |
|
not modelled |
18.7 |
9 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:N-6 DNA Methylase-like |
|
|
| 48 | c5ybbA_ |
|
not modelled |
18.5 |
17 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:type i restriction-modification system methyltransferase
PDBTitle: structural basis underlying complex assembly andconformational2 transition of the type i r-m system
|
|
|
| 49 | c1vzhB_ |
|
not modelled |
16.7 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:desulfoferrodoxin;
PDBTitle: structure of superoxide reductase bound to ferrocyanide and active2 site expansion upon x-ray induced photoreduction
|
|
|
| 50 | c3khkA_ |
|
not modelled |
15.1 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:type i restriction-modification system methylation subunit;
PDBTitle: crystal structure of type-i restriction-modification system2 methylation subunit (mm_0429) from methanosarchina mazei.
|
|
|
| 51 | c2waqG_ |
|
not modelled |
14.6 |
25 |
PDB header:transcription
Chain: G: PDB Molecule:dna-directed rna polymerase rpo8 subunit;
PDBTitle: the complete structure of the archaeal 13-subunit dna-directed rna2 polymerase
|
|
|
| 52 | d2ayxa2 |
|
not modelled |
14.2 |
21 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:RcsC linker domain-like |
|
|
| 53 | c2l3bA_ |
|
not modelled |
14.1 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein found in conjugate transposon;
PDBTitle: solution nmr structure of the bt_0084 lipoprotein from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr376
|
|
|
| 54 | d1jwya1 |
|
not modelled |
14.0 |
28 |
Fold:SH3-like barrel
Superfamily:Myosin S1 fragment, N-terminal domain
Family:Myosin S1 fragment, N-terminal domain |
|
|
| 55 | c3lkdB_ |
|
not modelled |
13.9 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:type i restriction-modification system methyltransferase
PDBTitle: crystal structure of the type i restriction-modification system2 methyltransferase subunit from streptococcus thermophilus, northeast3 structural genomics consortium target sur80
|
|
|
| 56 | c3ufbA_ |
|
not modelled |
13.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:type i restriction-modification system methyltransferase
PDBTitle: crystal structure of a modification subunit of a putative type i2 restriction enzyme from vibrio vulnificus yj016
|
|
|
| 57 | c2l7qA_ |
|
not modelled |
13.3 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein found in conjugate transposon;
PDBTitle: solution nmr structure of conjugate transposon protein bvu_1572(27-2 141) from bacteroides vulgatus, northeast structural genomics3 consortium target bvr155
|
|
|
| 58 | c3p9nA_ |
|
not modelled |
13.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:possible methyltransferase (methylase);
PDBTitle: rv2966c of m. tuberculosis is a rsmd-like methyltransferase
|
|
|
| 59 | c3j3bU_ |
|
not modelled |
12.7 |
7 |
PDB header:ribosome
Chain: U: PDB Molecule:60s ribosomal protein l22;
PDBTitle: structure of the human 60s ribosomal proteins
|
|
|
| 60 | c4qxlA_ |
|
not modelled |
12.2 |
20 |
PDB header:motor protein
Chain: A: PDB Molecule:flagellar protein flhe;
PDBTitle: crystal structure of flhe
|
|
|
| 61 | c5hfjF_ |
|
not modelled |
12.1 |
31 |
PDB header:dna binding protein
Chain: F: PDB Molecule:adenine specific dna methyltransferase (dpna);
PDBTitle: crystal structure of m1.hpyavi-sam complex
|
|
|
| 62 | c3j39U_ |
|
not modelled |
12.1 |
7 |
PDB header:ribosome
Chain: U: PDB Molecule:60s ribosomal protein l22;
PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
|
|
|
| 63 | c1aqjB_ |
|
not modelled |
12.0 |
21 |
PDB header:methyltransferase
Chain: B: PDB Molecule:adenine-n6-dna-methyltransferase taqi;
PDBTitle: structure of adenine-n6-dna-methyltransferase taqi
|
|
|
| 64 | c4lbaB_ |
|
not modelled |
11.5 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conjugative transposon lipoprotein;
PDBTitle: crystal structure of a conjugative transposon lipoprotein2 (bacegg_03088) from bacteroides eggerthii dsm 20697 at 1.70 a3 resolution
|
|
|
| 65 | d2okca1 |
|
not modelled |
11.3 |
22 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:N-6 DNA Methylase-like |
|
|
| 66 | c4zyeA_ |
|
not modelled |
10.8 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:methylated-dna--protein-cysteine methyltransferase;
PDBTitle: crystal structure of sulfolobus solfataricus o6-methylguanine2 methyltransferase
|
|
|
| 67 | c6h2uA_ |
|
not modelled |
10.8 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase-like protein 5;
PDBTitle: crystal structure of human mettl5-trmt112 complex, the 18s rrna2 m6a1832 methyltransferase at 1.6a resolution
|
|
|
| 68 | d2f8la1 |
|
not modelled |
10.6 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:N-6 DNA Methylase-like |
|
|
| 69 | c4qh0D_ |
|
not modelled |
10.3 |
9 |
PDB header:hydrolase
Chain: D: PDB Molecule:dna-entry nuclease (competence-specific nuclease);
PDBTitle: crystal structure of nuca from streptococcus agalactiae with magnesium2 ion bound
|
|
|
| 70 | c1xtyD_ |
|
not modelled |
10.1 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of sulfolobus solfataricus peptidyl-trna hydrolase
|
|
|
| 71 | c3ldgA_ |
|
not modelled |
9.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative uncharacterized protein smu.472;
PDBTitle: crystal structure of smu.472, a putative methyltransferase complexed2 with sah
|
|
|
| 72 | d1dkua1 |
|
not modelled |
9.8 |
17 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
|
|
| 73 | d1rlka_ |
|
not modelled |
9.7 |
15 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
|
|
| 74 | c2zv3E_ |
|
not modelled |
9.6 |
10 |
PDB header:hydrolase
Chain: E: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of project mj0051 from methanocaldococcus2 jannaschii dsm 2661
|
|
|
| 75 | c2vwtA_ |
|
not modelled |
9.5 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:yfau, 2-keto-3-deoxy sugar aldolase;
PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
|
|
|
| 76 | c1g38A_ |
|
not modelled |
9.5 |
21 |
PDB header:transferase/dna
Chain: A: PDB Molecule:modification methylase taqi;
PDBTitle: adenine-specific methyltransferase m. taq i/dna complex
|
|
|
| 77 | d1ne2a_ |
|
not modelled |
9.5 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Ta1320-like |
|
|
| 78 | c2g7hA_ |
|
not modelled |
9.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:methylated-dna--protein-cysteine
PDBTitle: structure of an o6-methylguanine dna methyltransferase from2 methanococcus jannaschii (mj1529)
|
|
|
| 79 | c4tv6A_ |
|
not modelled |
9.2 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxyglucarate aldolase;
PDBTitle: crystal structure of citrate synthase variant sbng e151q
|
|
|
| 80 | c6b3oB_ |
|
not modelled |
9.0 |
31 |
PDB header:viral protein
Chain: B: PDB Molecule:spike glycoprotein;
PDBTitle: tectonic conformational changes of a coronavirus spike glycoprotein2 promote membrane fusion
|
|
|
| 81 | c3hvzB_ |
|
not modelled |
8.8 |
7 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
|
|
|
| 82 | c5fgwA_ |
|
not modelled |
8.6 |
24 |
PDB header:metal binding, dna binding protein
Chain: A: PDB Molecule:extracellular streptodornase d;
PDBTitle: structure of sda1 nuclease with bound zinc ion
|
|
|
| 83 | c3owvA_ |
|
not modelled |
8.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:dna-entry nuclease;
PDBTitle: structural insights into catalytic and substrate binding mechanisms of2 the strategic enda nuclease from streptococcus pneumoniae
|
|
|
| 84 | d1wy7a1 |
|
not modelled |
8.4 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Ta1320-like |
|
|
| 85 | c5e72A_ |
|
not modelled |
8.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:n2, n2-dimethylguanosine trna methyltransferase;
PDBTitle: crystal structure of the archaeal trna m2g/m22g10 methyltransferase2 (atrm11) in complex with s-adenosyl-l-methionine (sam) from3 thermococcus kodakarensis
|
|
|
| 86 | d1xo8a_ |
|
not modelled |
8.3 |
14 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:LEA14-like
Family:LEA14-like |
|
|
| 87 | c5egpB_ |
|
not modelled |
8.3 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:ubie/coq5 family methyltransferase, putative;
PDBTitle: crystal structure of the s-methyltransferase tmta
|
|
|
| 88 | d1mgta1 |
|
not modelled |
7.8 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Methylated DNA-protein cysteine methyltransferase, C-terminal domain
Family:Methylated DNA-protein cysteine methyltransferase, C-terminal domain |
|
|
| 89 | d1bf2a2 |
|
not modelled |
7.7 |
10 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
|
|
| 90 | d2esra1 |
|
not modelled |
7.5 |
20 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
|
|
| 91 | d1ws6a1 |
|
not modelled |
7.5 |
27 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
|
|
| 92 | c3qz6A_ |
|
not modelled |
7.4 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:hpch/hpai aldolase;
PDBTitle: the crystal structure of hpch/hpai aldolase from desulfitobacterium2 hafniense dcb-2
|
|
|
| 93 | d2p92a1 |
|
not modelled |
7.4 |
16 |
Fold:eIF1-like
Superfamily:TM1457-like
Family:TM1457-like |
|
|
| 94 | c3cioA_ |
|
not modelled |
7.4 |
27 |
PDB header:signaling protein, transferase
Chain: A: PDB Molecule:tyrosine-protein kinase etk;
PDBTitle: the kinase domain of escherichia coli tyrosine kinase etk
|
|
|
| 95 | c2xh3B_ |
|
not modelled |
7.3 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:spd1 nuclease;
PDBTitle: extracellular nuclease
|
|
|
| 96 | c4he5A_ |
|
not modelled |
7.1 |
8 |
PDB header:unknown function
Chain: A: PDB Molecule:peptidase family u32;
PDBTitle: crystal structure of the selenomethionine variant of the c-terminal2 domain of geobacillus thermoleovorans putative u32 peptidase
|
|
|
| 97 | c2d3kA_ |
|
not modelled |
7.1 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: structural study on project id ph1539 from pyrococcus2 horikoshii ot3
|
|
|
| 98 | c2p35A_ |
|
not modelled |
7.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:trans-aconitate 2-methyltransferase;
PDBTitle: crystal structure of trans-aconitate methyltransferase from2 agrobacterium tumefaciens
|
|
|
| 99 | d2fpoa1 |
|
not modelled |
7.0 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
|
|
































































































































































































