| 1 | c3vsjB_
|
|
 |
100.0 |
15 |
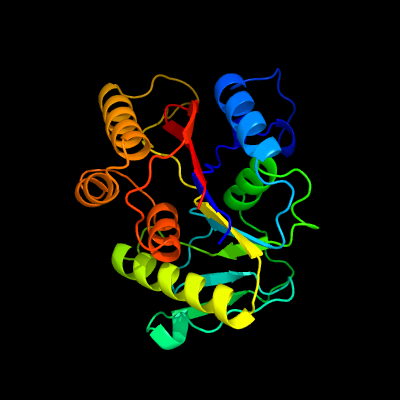
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-amino-5-chlorophenol 1,6-dioxygenase beta subunit;
PDBTitle: crystal structure of 1,6-apd (2-animophenol-1,6-dioxygenase) complexed2 with intermediate products
|
|
|
|
| 2 | c3vsjA_
|
|
 |
100.0 |
13 |
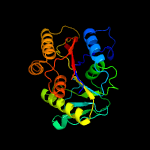
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit;
PDBTitle: crystal structure of 1,6-apd (2-animophenol-1,6-dioxygenase) complexed2 with intermediate products
|
|
|
|
| 3 | d1b4ub_
|
|
 |
100.0 |
12 |
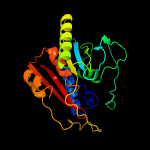
Fold:Phosphorylase/hydrolase-like
Superfamily:LigB-like
Family:LigB-like |
|
|
|
| 4 | c3wrbB_
|
|
 |
100.0 |
12 |
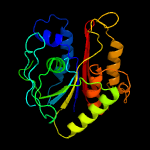
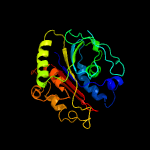
PDB header:oxidoreductase
Chain: B: PDB Molecule:gallate dioxygenase;
PDBTitle: crystal structure of the anaerobic h124f desb-gallate complex
|
|
|
|
| 5 | c3bd0D_
|
|
 |
99.9 |
15 |
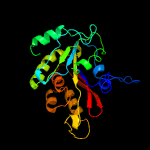
PDB header:peptide binding protein
Chain: D: PDB Molecule:protein memo1;
PDBTitle: crystal structure of memo, form ii
|
|
|
|
| 6 | d2pw6a1
|
|
 |
99.8 |
12 |
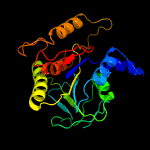
Fold:Phosphorylase/hydrolase-like
Superfamily:LigB-like
Family:LigB-like |
|
|
|
| 7 | d1a73a_
|
|
 |
22.8 |
30 |
Fold:His-Me finger endonucleases
Superfamily:His-Me finger endonucleases
Family:Intron-encoded homing endonucleases |
|
|
|
| 8 | c3wrwE_
|
|
 |
22.6 |
14 |
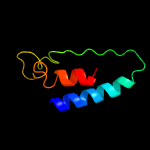
PDB header:transferase
Chain: E: PDB Molecule:tm-1 protein;
PDBTitle: crystal structure of the n-terminal domain of resistance protein
|
|
|
|
| 9 | c4uplC_
|
|
 |
19.1 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:sulfatase family protein;
PDBTitle: dimeric sulfatase spas2 from silicibacter pomeroyi
|
|
|
|
| 10 | d1y6va1
|
|
 |
11.8 |
18 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Alkaline phosphatase |
|
|
|
| 11 | c4s1nA_
|
|
 |
11.7 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from streptococcus pneumoniae tigr4
|
|
|
|
| 12 | c5c2gD_
|
|
 |
11.4 |
10 |
PDB header:lyase
Chain: D: PDB Molecule:form ii rubisco;
PDBTitle: gws1b rubisco: form ii rubisco derived from uncultivated2 gallionellacea species (cabp-bound).
|
|
|
|
| 13 | c2rusB_
|
|
 |
10.2 |
11 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:rubisco (ribulose-1,5-bisphosphate carboxylase(slash)
PDBTitle: crystal structure of the ternary complex of ribulose-1,5-bisphosphate2 carboxylase, mg(ii), and activator co2 at 2.3-angstroms resolution
|
|
|
|
| 14 | c9rubB_
|
|
 |
9.7 |
11 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:ribulose-1,5-bisphosphate carboxylase;
PDBTitle: crystal structure of activated ribulose-1,5-bisphosphate carboxylase2 complexed with its substrate, ribulose-1,5-bisphosphate
|
|
|
|
| 15 | d1ac5a_
|
|
 |
9.6 |
20 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Serine carboxypeptidase-like |
|
|
|
| 16 | d2ihta3
|
|
 |
9.4 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
|
|
|
| 17 | d5ruba1
|
|
 |
9.2 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
|
| 18 | c6b1vB_
|
|
 |
8.6 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:iota-carrageenan sulfatase;
PDBTitle: crystal structure of ps i-cgsb c78s in complex with i-neocarratetraose
|
|
|
|
| 19 | c1gxsC_
|
|
 |
7.8 |
36 |
PDB header:lyase
Chain: C: PDB Molecule:p-(s)-hydroxymandelonitrile lyase chain a;
PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
|
|
|
|
| 20 | c3p9xB_
|
|
 |
7.7 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
|
|
|
|
| 21 | c4az3A_ |
|
not modelled |
7.5 |
40 |
PDB header:hydrolase
Chain: A: PDB Molecule:lysosomal protective protein 32 kda chain;
PDBTitle: crystal structure of cathepsin a, complexed with 15a
|
|
|
| 22 | c1bcrA_ |
|
not modelled |
7.3 |
36 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:serine carboxypeptidase ii;
PDBTitle: complex of the wheat serine carboxypeptidase, cpdw-ii, with the2 microbial peptide aldehyde inhibitor, antipain, and arginine at room3 temperature
|
|
|
| 23 | c3uebD_ |
|
not modelled |
7.2 |
9 |
PDB header:unknown function
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of ton_0450 from thermococcus onnurineus na1
|
|
|
| 24 | c3a52A_ |
|
not modelled |
7.2 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:cold-active alkaline phosphatase;
PDBTitle: crystal structure of cold-active alkailne phosphatase from2 psychrophile shewanella sp.
|
|
|
| 25 | d1wpxa1 |
|
not modelled |
6.9 |
27 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Serine carboxypeptidase-like |
|
|
| 26 | c5macD_ |
|
not modelled |
6.8 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:ribulose-1,5-bisphosphate carboxylase-oxygenase type iii;
PDBTitle: crystal structure of decameric methanococcoides burtonii rubisco2 complexed with 2-carboxyarabinitol bisphosphate
|
|
|
| 27 | c2x9qA_ |
|
not modelled |
6.5 |
8 |
PDB header:ligase
Chain: A: PDB Molecule:cyclodipeptide synthetase;
PDBTitle: structure of the mycobacterium tuberculosis protein, rv2275,2 demonstrates that cyclodipeptide synthetases are related3 to type i trna-synthetases.
|
|
|
| 28 | d1ovma3 |
|
not modelled |
6.4 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
|
|
| 29 | c2qygC_ |
|
not modelled |
6.0 |
11 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
|
|
| 30 | d1rbla1 |
|
not modelled |
5.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 31 | d1q5qa_ |
|
not modelled |
5.2 |
15 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Proteasome subunits |
|
|
| 32 | c1q5rD_ |
|
not modelled |
5.2 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:proteasome alpha-type subunit 1;
PDBTitle: the rhodococcus 20s proteasome with unprocessed pro-peptides
|
|
|
| 33 | d1nvpd1 |
|
not modelled |
5.2 |
23 |
Fold:Transcription factor IIA (TFIIA), alpha-helical domain
Superfamily:Transcription factor IIA (TFIIA), alpha-helical domain
Family:Transcription factor IIA (TFIIA), alpha-helical domain |
|
|
| 34 | d1geha1 |
|
not modelled |
5.1 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 35 | d2djia3 |
|
not modelled |
5.1 |
27 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
|
|
| 36 | c2rm8A_ |
|
not modelled |
5.1 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:sensory rhodopsin ii transducer;
PDBTitle: the solution structure of phototactic transducer protein2 htrii linker region from natronomonas pharaonis
|
|
|