| 1 |
|
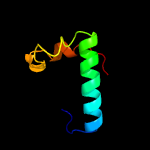
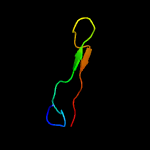
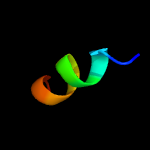
PDB 2knu chain A
Region: 1 - 8
Aligned: 8
Modelled: 8
Confidence: 50.4%
Identity: 75%
PDB header:membrane protein
Chain: A: PDB Molecule:genome polyprotein;
PDBTitle: solution structure of the transmembrane proximal region of2 the hepatis c virus e1 glycoprotein
Phyre2
| 2 |
|
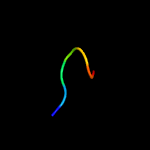
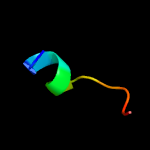
PDB 6atk chain E
Region: 53 - 70
Aligned: 18
Modelled: 18
Confidence: 20.8%
Identity: 33%
PDB header:hydrolase/viral protein
Chain: E: PDB Molecule:spike glycoprotein;
PDBTitle: crystal structure of the human coronavirus 229e spike protein receptor2 binding domain in complex with human aminopeptidase n
Phyre2
| 3 |
|
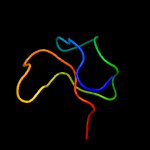
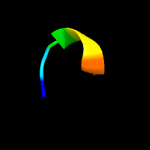
PDB 1vw4 chain G
Region: 34 - 42
Aligned: 9
Modelled: 9
Confidence: 14.1%
Identity: 67%
PDB header:ribosome
Chain: G: PDB Molecule:54s ribosomal protein l50, mitochondrial;
PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
Phyre2
| 4 |
|
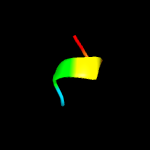
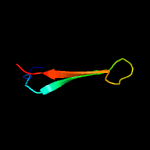
PDB 3kk4 chain B
Region: 109 - 158
Aligned: 50
Modelled: 50
Confidence: 13.2%
Identity: 28%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein bp1543;
PDBTitle: uncharacterized protein bp1543 from bordetella pertussis tohama i
Phyre2
| 5 |
|
PDB 2m7t chain A
Region: 83 - 91
Aligned: 9
Modelled: 9
Confidence: 11.9%
Identity: 67%
PDB header:protein binding
Chain: A: PDB Molecule:cystine knot protein 2.5d;
PDBTitle: solution nmr structure of engineered cystine knot protein 2.5d
Phyre2
| 6 |
|
PDB 5a4h chain A
Region: 85 - 91
Aligned: 7
Modelled: 7
Confidence: 8.6%
Identity: 71%
PDB header:transferase
Chain: A: PDB Molecule:1-acylglycerol-3-phosphate o-acyltransferase abhd5;
PDBTitle: solution structure of the lipid droplet anchoring peptide2 of cgi-58 bound to dpc micelles
Phyre2
| 7 |
|
PDB 2ahq chain A
Region: 107 - 113
Aligned: 7
Modelled: 7
Confidence: 8.2%
Identity: 57%
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma factor rpon;
PDBTitle: solution structure of the c-terminal rpon domain of sigma-2 54 from aquifex aeolicus
Phyre2
| 8 |
|
PDB 1koz chain A
Region: 65 - 95
Aligned: 31
Modelled: 31
Confidence: 7.9%
Identity: 35%
PDB header:toxin
Chain: A: PDB Molecule:voltage-dependent channel inhibitor;
PDBTitle: solution structure of omega-grammotoxin sia
Phyre2
| 9 |
|
PDB 3nct chain C
Region: 25 - 31
Aligned: 7
Modelled: 7
Confidence: 7.7%
Identity: 71%
PDB header:dna binding protein, chaperone
Chain: C: PDB Molecule:protein psib;
PDBTitle: x-ray crystal structure of the bacterial conjugation factor psib, a2 negative regulator of reca
Phyre2
| 10 |
|
PDB 1sb7 chain A
Region: 61 - 66
Aligned: 6
Modelled: 6
Confidence: 7.2%
Identity: 67%
PDB header:lyase
Chain: A: PDB Molecule:trna pseudouridine synthase d;
PDBTitle: crystal structure of the e.coli pseudouridine synthase trud
Phyre2
| 11 |
|
PDB 2kzk chain A
Region: 45 - 72
Aligned: 28
Modelled: 28
Confidence: 7.2%
Identity: 21%
PDB header:protein transport
Chain: A: PDB Molecule:uncharacterized protein yol083w;
PDBTitle: solution structure of alpha-mannosidase binding domain of atg34
Phyre2
| 12 |
|
PDB 2vqz chain B
Region: 51 - 72
Aligned: 22
Modelled: 22
Confidence: 6.8%
Identity: 14%
PDB header:transcription
Chain: B: PDB Molecule:polymerase basic protein 2;
PDBTitle: structure of the cap-binding domain of influenza virus2 polymerase subunit pb2 with bound m7gtp
Phyre2
| 13 |
|
PDB 6cfz chain J
Region: 24 - 35
Aligned: 12
Modelled: 12
Confidence: 6.4%
Identity: 25%
PDB header:nuclear protein
Chain: J: PDB Molecule:spc34;
PDBTitle: structure of the dash/dam1 complex shows its role at the yeast2 kinetochore-microtubule interface
Phyre2
| 14 |
|
PDB 1szw chain A
Region: 61 - 66
Aligned: 6
Modelled: 6
Confidence: 6.0%
Identity: 67%
Fold: Pseudouridine synthase
Superfamily: Pseudouridine synthase
Family: tRNA pseudouridine synthase TruD
Phyre2
| 15 |
|
PDB 4gu8 chain G
Region: 89 - 97
Aligned: 9
Modelled: 9
Confidence: 6.0%
Identity: 78%
PDB header:sugar binding protein
Chain: G: PDB Molecule:burkholderia oklahomensis agglutinin (boa);
PDBTitle: crystal structure of burkholderia oklahomensis agglutinin (boa)
Phyre2
| 16 |
|
PDB 2kzb chain A
Region: 45 - 72
Aligned: 28
Modelled: 28
Confidence: 6.0%
Identity: 29%
PDB header:protein transport
Chain: A: PDB Molecule:autophagy-related protein 19;
PDBTitle: solution structure of alpha-mannosidase binding domain of atg19
Phyre2
| 17 |
|
PDB 1bkv chain A
Region: 128 - 137
Aligned: 10
Modelled: 10
Confidence: 5.8%
Identity: 70%
PDB header:structural protein
Chain: A: PDB Molecule:t3-785;
PDBTitle: collagen
Phyre2
| 18 |
|
PDB 2lr3 chain A
Region: 64 - 75
Aligned: 12
Modelled: 12
Confidence: 5.8%
Identity: 42%
PDB header:antifungal protein
Chain: A: PDB Molecule:defensin;
PDBTitle: solution structure of the anti-fungal defensin def4 (mtr_8g070770)2 from medicago truncatula (barrel clover)
Phyre2
| 19 |
|
PDB 5zgh chain I
Region: 117 - 128
Aligned: 12
Modelled: 12
Confidence: 5.5%
Identity: 67%
PDB header:photosynthesis
Chain: I: PDB Molecule:psai;
PDBTitle: cryo-em structure of the red algal psi-lhcr
Phyre2
| 20 |
|
PDB 1bkv chain B
Region: 128 - 137
Aligned: 10
Modelled: 10
Confidence: 5.3%
Identity: 70%
PDB header:structural protein
Chain: B: PDB Molecule:t3-785;
PDBTitle: collagen
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|