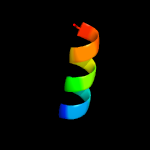
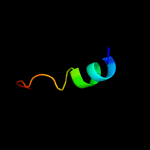
1 c6jx3B_
51.6
31
PDB header: peptide binding proteinChain: B: PDB Molecule: tfub1;PDBTitle: lasso peptide synthetase b1 complexed with the leader peptide
2 d1ecia_
43.6
42
Fold: Ectatomin subunitsSuperfamily: Ectatomin subunitsFamily: Ectatomin subunits
3 c5v1uB_
43.2
28
PDB header: protein bindingChain: B: PDB Molecule: tbib1;PDBTitle: tbib1 in complex with the tbia(beta) leader peptide
4 c4bx9C_
40.1
25
PDB header: protein transportChain: C: PDB Molecule: vacuolar protein sorting-associated protein 16 homolog;PDBTitle: human vps33a in complex with a fragment of human vps16
5 c6fkgC_
39.6
16
PDB header: toxinChain: C: PDB Molecule: rv1990c (mbca);PDBTitle: crystal structure of the m.tuberculosis mbct-mbca toxin-antitoxin2 complex.
6 c2nydB_
29.1
8
PDB header: unknown functionChain: B: PDB Molecule: upf0135 protein sa1388;PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
7 c2p9xB_
25.6
28
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein ph0832;PDBTitle: crystal structure of ph0832 from pyrococcus horikoshii ot3
8 c3ctdB_
22.0
32
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative atpase, aaa family;PDBTitle: crystal structure of a putative aaa family atpase from prochlorococcus2 marinus subsp. pastoris
9 c3g2bA_
21.1
20
PDB header: biosynthetic proteinChain: A: PDB Molecule: coenzyme pqq synthesis protein d;PDBTitle: crystal structure of pqqd from xanthomonas campestris
10 c4na3A_
20.6
30
PDB header: transferaseChain: A: PDB Molecule: polyketide synthase pksj;PDBTitle: crystal structure of the second ketosynthase from the bacillaene2 polyketide synthase bound to a hexanoyl substrate mimic
11 d3ctda1
18.1
32
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like
12 c6gw6B_
17.5
20
PDB header: toxinChain: B: PDB Molecule: xre antitoxin;PDBTitle: structure of the pseudomonas putida res-xre toxin-antitoxin complex
13 c4z37A_
17.1
23
PDB header: transferaseChain: A: PDB Molecule: putative mixed polyketide synthase/non-ribosomal peptidePDBTitle: structure of the ketosynthase of module 2 of c0zgq5 (trans-at pks)2 from brevibacillus brevis
14 c3twkB_
16.0
17
PDB header: hydrolaseChain: B: PDB Molecule: formamidopyrimidine-dna glycosylase 1;PDBTitle: crystal structure of arabidopsis thaliana fpg
15 c5erbB_
15.9
31
PDB header: transferaseChain: B: PDB Molecule: polyketide synthase;PDBTitle: ketosynthase from module 5 of the bacillaene synthase from bacillus2 amyloliquefaciens fzb42
16 c4tl2A_
15.3
36
PDB header: transferaseChain: A: PDB Molecule: at-less polyketide synthase;PDBTitle: crystal structure of ketosynthase domain from mgsf from streptomyces2 platensis
17 c4tktA_
15.1
26
PDB header: transferaseChain: A: PDB Molecule: at-less polyketide synthase;PDBTitle: streptomyces platensis isomigrastatin ketosynthase domain mgsf ks6
18 c3p9aI_
14.5
38
PDB header: dna binding proteinChain: I: PDB Molecule: dna-packaging protein gp3;PDBTitle: an atomic view of the nonameric small terminase subunit of2 bacteriophage p22
19 d3bgea1
14.0
37
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like
20 c3ismC_
13.5
21
PDB header: hydrolase inhibitor/hydrolaseChain: C: PDB Molecule: cg4930;PDBTitle: crystal structure of the endog/endogi complex: mechanism of endog2 inhibition
21 c4qyrA_
not modelled
13.4
29
PDB header: transferaseChain: A: PDB Molecule: at-less polyketide synthase;PDBTitle: streptomyces platensis isomigrastatin ketosynthase domain mgse ks3
22 c4b3nA_
not modelled
13.3
19
PDB header: sugar binding protein/ligaseChain: A: PDB Molecule: maltose-binding periplasmic protein, tripartite motif-PDBTitle: crystal structure of rhesus trim5alpha pry/spry domain
23 d1rd5a_
not modelled
12.9
26
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes
24 c5e5nD_
not modelled
12.9
29
PDB header: hydrolaseChain: D: PDB Molecule: polyketide synthase pksl;PDBTitle: ketosynthase from module 6 of the bacillaene synthase from bacillus2 subtilis 168 (c167s mutant, crystal form 1)
25 c3qyxD_
not modelled
12.7
11
PDB header: transcription/dnaChain: D: PDB Molecule: esx-1 secretion-associated regulator espr;PDBTitle: crystal structure of mycobacterium tuberculosis espr in complex with a2 small dna fragment
26 d1ee8a1
not modelled
12.5
13
Fold: S13-like H2TH domainSuperfamily: S13-like H2TH domainFamily: Middle domain of MutM-like DNA repair proteins
27 c5e5nB_
not modelled
12.0
36
PDB header: hydrolaseChain: B: PDB Molecule: polyketide synthase pksl;PDBTitle: ketosynthase from module 6 of the bacillaene synthase from bacillus2 subtilis 168 (c167s mutant, crystal form 1)
28 c1no3A_
not modelled
11.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: lipoxygenase-3;PDBTitle: refined structure of soybean lipoxygenase-3 with 4-nitrocatechol at2 2.15 angstrom resolution
29 c5b88A_
not modelled
11.5
30
PDB header: rna binding proteinChain: A: PDB Molecule: atp-dependent rna helicase dead;PDBTitle: rrm-like domain of dead-box protein, csda
30 d1rrha1
not modelled
11.1
18
Fold: LipoxigenaseSuperfamily: LipoxigenaseFamily: Plant lipoxigenases
31 c3iz5w_
not modelled
10.5
16
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein l22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
32 c3izcw_
not modelled
10.4
32
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein rpl22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
33 c6fahD_
not modelled
10.1
15
PDB header: flavoproteinChain: D: PDB Molecule: caffeyl-coa reductase-etf complex subunit carc;PDBTitle: molecular basis of the flavin-based electron-bifurcating caffeyl-coa2 reductase reaction
34 c5gxtA_
not modelled
9.4
19
PDB header: protein transportChain: A: PDB Molecule: maltose-binding periplasmic protein,pigg;PDBTitle: crystal structure of pigg
35 c4a5uA_
not modelled
9.4
21
PDB header: transferase/rna binding proteinChain: A: PDB Molecule: rna replicase polyprotein;PDBTitle: turnip yellow mosaic virus proteinase and escherichia coli 30s2 ribosomal s15
36 c4opeD_
not modelled
9.3
34
PDB header: ligase, transferaseChain: D: PDB Molecule: nrps/pks;PDBTitle: streptomcyes albus ja3453 oxazolomycin ketosynthase domain ozmh ks7
37 c4wviA_
not modelled
8.6
26
PDB header: hydrolaseChain: A: PDB Molecule: maltose-binding periplasmic protein,signal peptidase ib;PDBTitle: crystal structure of the type-i signal peptidase from staphylococcus2 aureus (spsb) in complex with a substrate peptide (pep2).
38 c3csgA_
not modelled
8.6
19
PDB header: de novo protein, sugar binding proteinChain: A: PDB Molecule: maltose-binding protein monobody ys1 fusion;PDBTitle: crystal structure of monobody ys1(mbp-74)/maltose binding protein2 fusion complex
39 c4wkyB_
not modelled
8.4
17
PDB header: transferaseChain: B: PDB Molecule: beta-ketoacyl synthase;PDBTitle: streptomcyes albus ja3453 oxazolomycin ketosynthase domain ozmn ks2
40 c5sxyA_
not modelled
8.2
8
PDB header: chaperoneChain: A: PDB Molecule: bifunctional coenzyme pqq synthesis protein c/d;PDBTitle: the solution nmr structure for the pqqd truncation of methylobacterium2 extorquens pqqcd representing a functional and stand-alone3 ribosomally synthesized and post-translational modified (ripp)4 recognition element (rre)
41 d2proc1
not modelled
8.1
27
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Alpha-lytic protease prodomainFamily: Alpha-lytic protease prodomain
42 c3hbmA_
not modelled
7.9
5
PDB header: hydrolaseChain: A: PDB Molecule: udp-sugar hydrolase;PDBTitle: crystal structure of pseg from campylobacter jejuni
43 d3bula1
not modelled
7.6
21
Fold: Methionine synthase domain-likeSuperfamily: Methionine synthase domainFamily: Methionine synthase domain
44 d1k82a1
not modelled
7.5
13
Fold: S13-like H2TH domainSuperfamily: S13-like H2TH domainFamily: Middle domain of MutM-like DNA repair proteins
45 c3p14C_
not modelled
7.5
25
PDB header: isomeraseChain: C: PDB Molecule: l-rhamnose isomerase;PDBTitle: crystal structure of l-rhamnose isomerase with a novel high thermo-2 stability from bacillus halodurans
46 c2qw6A_
not modelled
7.5
31
PDB header: hydrolaseChain: A: PDB Molecule: aaa atpase, central region;PDBTitle: crystal structure of the c-terminal domain of an aaa atpase from2 enterococcus faecium do
47 d2qw6a1
not modelled
7.5
31
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like
48 c3oaiB_
not modelled
7.4
19
PDB header: membrane protein, cell adhesionChain: B: PDB Molecule: maltose-binding periplasmic protein, myelin protein p0;PDBTitle: crystal structure of the extra-cellular domain of human myelin protein2 zero
49 c3msqC_
not modelled
7.4
7
PDB header: biosynthetic proteinChain: C: PDB Molecule: putative ubiquinone biosynthesis protein;PDBTitle: crystal structure of a putative ubiquinone biosynthesis protein2 (npun02000094) from nostoc punctiforme pcc 73102 at 2.85 a resolution
50 c6itwA_
not modelled
7.1
11
PDB header: antitoxinChain: A: PDB Molecule: type vi immunity protein atu4351;PDBTitle: crystal structure of atu4351 from agrobacterium tumefaciens
51 d2r9ga1
not modelled
7.0
26
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like
52 d1d8wa_
not modelled
6.9
30
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: L-rhamnose isomerase
53 c6bwqB_
not modelled
6.8
25
PDB header: metal binding proteinChain: B: PDB Molecule: pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickelPDBTitle: larc2, the c-terminal domain of a cyclometallase involved in the2 synthesis of the npn cofactor of lactate racemase, in complex with3 mnctp
54 c1ty4D_
not modelled
6.7
63
PDB header: apoptosisChain: D: PDB Molecule: egg laying defective egl-1, programmed cell deathPDBTitle: crystal structure of a ced-9/egl-1 complex
55 c2joiA_
not modelled
6.7
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ta0095;PDBTitle: nmr solution structure of hypothetical protein ta0095 from2 thermoplasma acidophilum
56 d1nmpa_
not modelled
6.7
18
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like
57 c3gx4X_
not modelled
6.6
40
PDB header: dna binding protein/dnaChain: X: PDB Molecule: alkyltransferase-like protein 1;PDBTitle: crystal structure analysis of s. pombe atl in complex with dna
58 d1mwwa_
not modelled
6.6
17
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: Hypothetical protein HI1388.1
59 d1u0bb1
not modelled
6.5
25
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
60 c1ty4C_
not modelled
6.4
63
PDB header: apoptosisChain: C: PDB Molecule: egg laying defective egl-1, programmed cell deathPDBTitle: crystal structure of a ced-9/egl-1 complex
61 c4kmoB_
not modelled
6.3
17
PDB header: transport proteinChain: B: PDB Molecule: putative vacuolar protein sorting-associated protein;PDBTitle: crystal structure of the vps33-vps16 hops subcomplex from chaetomium2 thermophilum
62 d1nvma1
not modelled
6.3
16
Fold: RuvA C-terminal domain-likeSuperfamily: post-HMGL domain-likeFamily: DmpG/LeuA communication domain-like
63 c3k1lA_
not modelled
6.2
18
PDB header: ligaseChain: A: PDB Molecule: fancl;PDBTitle: crystal structure of fancl
64 c3zqsB_
not modelled
6.1
18
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase fancl;PDBTitle: human fancl central domain
65 c2gx8B_
not modelled
6.0
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: nif3-related protein;PDBTitle: the crystal structure of bacillus cereus protein related to nif3
66 c5vglA_
not modelled
6.0
16
PDB header: isomeraseChain: A: PDB Molecule: lachrymatory-factor synthase;PDBTitle: crystal structure of lachrymatory factor synthase from allium cepa
67 d1sr9a1
not modelled
6.0
29
Fold: RuvA C-terminal domain-likeSuperfamily: post-HMGL domain-likeFamily: DmpG/LeuA communication domain-like
68 d3proc1
not modelled
5.9
19
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Alpha-lytic protease prodomainFamily: Alpha-lytic protease prodomain
69 c2ix4B_
not modelled
5.9
24
PDB header: transferaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase;PDBTitle: arabidopsis thaliana mitochondrial beta-ketoacyl acp synthase hexanoic2 acid complex
70 c6d0hB_
not modelled
5.8
18
PDB header: toxinChain: B: PDB Molecule: pars: cog5642 (duf2384) antitoxin;PDBTitle: part: prs adp-ribosylating toxin bound to cognate antitoxin pars
71 c3c0vC_
not modelled
5.8
37
PDB header: plant proteinChain: C: PDB Molecule: cytokinin-specific binding protein;PDBTitle: crystal structure of cytokinin-specific binding protein in complex2 with cytokinin and ta6br12
72 c4kegA_
not modelled
5.7
21
PDB header: lipid binding proteinChain: A: PDB Molecule: maltose-binding periplasmic/palate lung and nasalPDBTitle: crystal structure of mbp fused human splunc1
73 c1dvaX_
not modelled
5.5
90
PDB header: hydrolase/hydrolase inhibitorChain: X: PDB Molecule: peptide e-76;PDBTitle: crystal structure of the complex between the peptide exosite inhibitor2 e-76 and coagulation factor viia
74 c1dvaY_
not modelled
5.5
90
PDB header: hydrolase/hydrolase inhibitorChain: Y: PDB Molecule: peptide e-76;PDBTitle: crystal structure of the complex between the peptide exosite inhibitor2 e-76 and coagulation factor viia
75 c3j61G_
not modelled
5.2
43
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein l8e;PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
76 c5elpA_
not modelled
5.1
6
PDB header: hydrolaseChain: A: PDB Molecule: nrps/pks protein;PDBTitle: ketosynthase from module 1 of the bacillaene synthase from bacillus2 amyloliquefaciens fzb42
77 c1r7gA_
not modelled
5.0
44
PDB header: membrane proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: nmr structure of the membrane anchor domain (1-31) of the2 nonstructural protein 5a (ns5a) of hepatitis c virus3 (minimized average structure, sample in 100mm dpc)