| 1 |
|
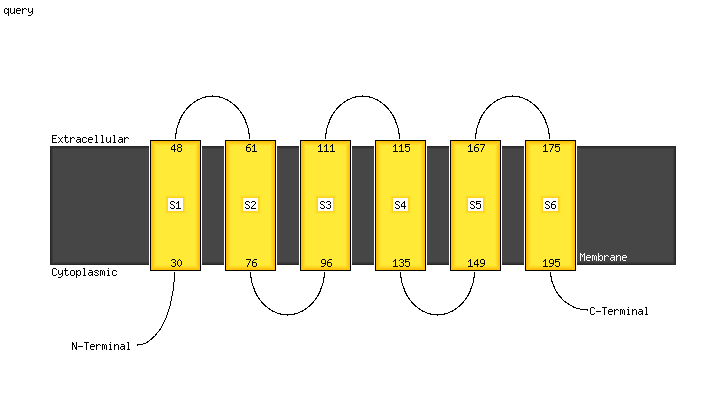
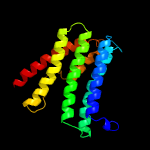
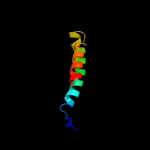
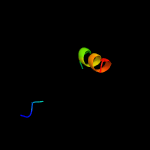
PDB 6h59 chain B
Region: 25 - 208
Aligned: 179
Modelled: 184
Confidence: 100.0%
Identity: 20%
PDB header:transferase
Chain: B: PDB Molecule:cdp-diacylglycerol--inositol 3-phosphatidyltransferase;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphatidylinositol2 phosphate synthase (pgsa1) with cdp-dag bound
Phyre2
| 2 |
|
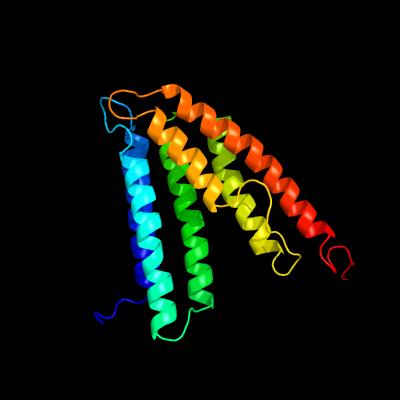
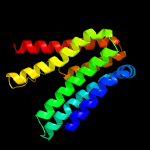
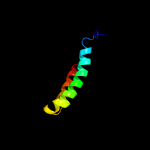
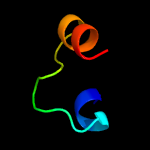
PDB 5d92 chain B
Region: 30 - 203
Aligned: 168
Modelled: 174
Confidence: 99.9%
Identity: 18%
PDB header:membrane protein
Chain: B: PDB Molecule:af2299 protein,phosphatidylinositol synthase;
PDBTitle: structure of a phosphatidylinositolphosphate (pip) synthase from2 renibacterium salmoninarum
Phyre2
| 3 |
|
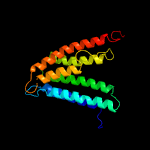
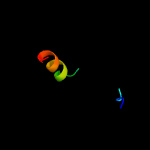
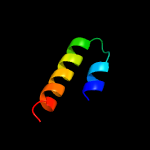
PDB 4o6m chain A
Region: 22 - 199
Aligned: 170
Modelled: 178
Confidence: 99.9%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:af2299, a cdp-alcohol phosphotransferase;
PDBTitle: structure of af2299, a cdp-alcohol phosphotransferase (cmp-bound)
Phyre2
| 4 |
|
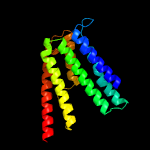
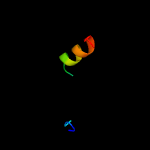
PDB 4mnd chain A
Region: 30 - 198
Aligned: 162
Modelled: 169
Confidence: 99.9%
Identity: 17%
PDB header:transferase
Chain: A: PDB Molecule:ctp l-myo-inositol-1-phosphate cytidylyltransferase/cdp-l-
PDBTitle: crystal structure of archaeoglobus fulgidus ipct-dipps bifunctional2 membrane protein
Phyre2
| 5 |
|
PDB 2voy chain G
Region: 84 - 112
Aligned: 29
Modelled: 29
Confidence: 32.5%
Identity: 14%
PDB header:hydrolase
Chain: G: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium atpase 1;
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 6 |
|
PDB 4i0x chain J
Region: 72 - 103
Aligned: 32
Modelled: 32
Confidence: 24.8%
Identity: 13%
PDB header:structural genomics, unknown function
Chain: J: PDB Molecule:esat-6-like protein mab_3113;
PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
Phyre2
| 7 |
|
PDB 6mjb chain C
Region: 81 - 98
Aligned: 18
Modelled: 18
Confidence: 23.3%
Identity: 17%
PDB header:cell cycle
Chain: C: PDB Molecule:kinetochore-associated protein dsn1;
PDBTitle: structure of candida glabrata csm1:dsn1(14-72) complex
Phyre2
| 8 |
|
PDB 2rkh chain A
Region: 67 - 107
Aligned: 41
Modelled: 41
Confidence: 21.9%
Identity: 17%
PDB header:transcription
Chain: A: PDB Molecule:putative apha-like transcription factor;
PDBTitle: crystal structure of a putative apha-like transcription factor2 (zp_00208345.1) from magnetospirillum magnetotacticum ms-1 at 2.00 a3 resolution
Phyre2
| 9 |
|
PDB 2le7 chain A
Region: 78 - 91
Aligned: 14
Modelled: 14
Confidence: 13.4%
Identity: 36%
PDB header:transport protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h member 2;
PDBTitle: solution nmr structure of the s4s5 linker of herg potassium channel
Phyre2
| 10 |
|
PDB 2mpn chain B
Region: 23 - 75
Aligned: 53
Modelled: 53
Confidence: 8.2%
Identity: 8%
PDB header:membrane protein
Chain: B: PDB Molecule:inner membrane protein ygap;
PDBTitle: 3d nmr structure of the transmembrane domain of the full-length inner2 membrane protein ygap from escherichia coli
Phyre2
| 11 |
|
PDB 2mpn chain A
Region: 23 - 75
Aligned: 53
Modelled: 53
Confidence: 8.2%
Identity: 8%
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein ygap;
PDBTitle: 3d nmr structure of the transmembrane domain of the full-length inner2 membrane protein ygap from escherichia coli
Phyre2
| 12 |
|
PDB 4n3x chain D
Region: 71 - 82
Aligned: 11
Modelled: 12
Confidence: 7.0%
Identity: 45%
PDB header:endocytosis
Chain: D: PDB Molecule:rab5 gdp/gtp exchange factor;
PDBTitle: crystal structure of rabex-5 cc domain
Phyre2
| 13 |
|
PDB 6mje chain B
Region: 82 - 98
Aligned: 17
Modelled: 17
Confidence: 5.9%
Identity: 18%
PDB header:cell cycle
Chain: B: PDB Molecule:dsn1p;
PDBTitle: structure of candida glabrata csm1: s. cerevisiae dsn1 complex
Phyre2
| 14 |
|
PDB 6mje chain H
Region: 82 - 98
Aligned: 17
Modelled: 17
Confidence: 5.9%
Identity: 18%
PDB header:cell cycle
Chain: H: PDB Molecule:dsn1p;
PDBTitle: structure of candida glabrata csm1: s. cerevisiae dsn1 complex
Phyre2
| 15 |
|
PDB 6mje chain F
Region: 82 - 98
Aligned: 17
Modelled: 17
Confidence: 5.9%
Identity: 18%
PDB header:cell cycle
Chain: F: PDB Molecule:dsn1p;
PDBTitle: structure of candida glabrata csm1: s. cerevisiae dsn1 complex
Phyre2
| 16 |
|
PDB 6mje chain D
Region: 82 - 98
Aligned: 17
Modelled: 17
Confidence: 5.9%
Identity: 18%
PDB header:cell cycle
Chain: D: PDB Molecule:dsn1p;
PDBTitle: structure of candida glabrata csm1: s. cerevisiae dsn1 complex
Phyre2
| 17 |
|
PDB 2cx6 chain A domain 1
Region: 78 - 101
Aligned: 24
Modelled: 23
Confidence: 5.8%
Identity: 17%
Fold: Barstar-like
Superfamily: Barstar-related
Family: Barstar-related
Phyre2
| 18 |
|
PDB 4arp chain B
Region: 67 - 96
Aligned: 30
Modelled: 30
Confidence: 5.3%
Identity: 13%
PDB header:hydrolase
Chain: B: PDB Molecule:pesticin;
PDBTitle: structure of the inactive pesticin e178a mutant
Phyre2