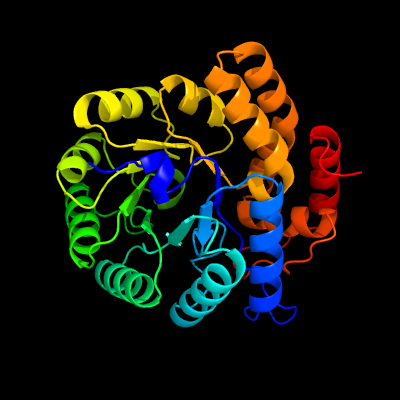
| 1 | d1xxxa1
|
|
 |
100.0 |
100 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
|
| 2 | c3cprB_
|
|
 |
100.0 |
58 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
|
|
|
| 3 | c3g0sA_
|
|
 |
100.0 |
33 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
|
|
|
|
| 4 | c3fluD_
|
|
 |
100.0 |
36 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
|
|
|
| 5 | c6mqhA_
|
|
 |
100.0 |
35 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate synthase (htpa2 synthase) from burkholderia mallei
|
|
|
|
| 6 | c3bi8A_
|
|
 |
100.0 |
35 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
|
|
|
| 7 | d2a6na1
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
|
| 8 | c3pueA_
|
|
 |
100.0 |
34 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
|
|
|
| 9 | c5ktlA_
|
|
 |
100.0 |
44 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
|
|
|
|
| 10 | c3noeA_
|
|
 |
100.0 |
36 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
|
|
|
| 11 | c2r8wB_
|
|
 |
100.0 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
|
|
|
| 12 | c3si9B_
|
|
 |
100.0 |
33 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
|
|
|
| 13 | c4n4qD_
|
|
 |
100.0 |
26 |
PDB header:lyase
Chain: D: PDB Molecule:acylneuraminate lyase;
PDBTitle: crystal structure of n-acetylneuraminate lyase from mycoplasma2 synoviae, crystal form ii
|
|
|
|
| 14 | d1f74a_
|
|
 |
100.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
|
| 15 | d1xkya1
|
|
 |
100.0 |
41 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
|
| 16 | c5afdA_
|
|
 |
100.0 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: native structure of n-acetylneuramininate lyase (sialic acid aldolase)2 from aliivibrio salmonicida
|
|
|
|
| 17 | c4ah7C_
|
|
 |
100.0 |
24 |
PDB header:lyase
Chain: C: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: structure of wild type stapylococcus aureus n-acetylneuraminic acid2 lyase in complex with pyruvate
|
|
|
|
| 18 | c2yxgD_
|
|
 |
100.0 |
34 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
|
|
|
| 19 | c5ud6B_
|
|
 |
100.0 |
40 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dhdps from cyanidioschyzon merolae with lysine2 bound
|
|
|
|
| 20 | c3eb2A_
|
|
 |
100.0 |
27 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
|
|
|
| 21 | c3s5oA_ |
|
not modelled |
100.0 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
|
|
| 22 | c4xkyC_ |
|
not modelled |
100.0 |
24 |
PDB header:lyase
Chain: C: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from the commensal bacterium2 bacteroides thetaiotaomicron at 2.1 a resolution
|
|
|
| 23 | d1hl2a_ |
|
not modelled |
100.0 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 24 | c2rfgB_ |
|
not modelled |
100.0 |
35 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
|
|
| 25 | c3h5dD_ |
|
not modelled |
100.0 |
36 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from drug-resistant streptococcus2 pneumoniae
|
|
|
| 26 | c4icnB_ |
|
not modelled |
100.0 |
35 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from shewanella benthica
|
|
|
| 27 | d1o5ka_ |
|
not modelled |
100.0 |
34 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 28 | c2ehhE_ |
|
not modelled |
100.0 |
34 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
|
|
| 29 | c3lciA_ |
|
not modelled |
100.0 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
|
|
| 30 | c2vc6A_ |
|
not modelled |
100.0 |
34 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
|
|
| 31 | c3na8A_ |
|
not modelled |
100.0 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
|
|
| 32 | c6h4eB_ |
|
not modelled |
100.0 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:putative n-acetylneuraminate lyase;
PDBTitle: proteus mirabilis n-acetylneuraminate lyase
|
|
|
| 33 | c4i7vD_ |
|
not modelled |
100.0 |
35 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: agrobacterium tumefaciens dhdps with pyruvate
|
|
|
| 34 | c4nq1B_ |
|
not modelled |
100.0 |
31 |
PDB header:lyase
Chain: B: PDB Molecule:4-hydroxy-tetrahydrodipicolinate synthase;
PDBTitle: legionella pneumophila dihydrodipicolinate synthase with first2 substrate pyruvate bound in the active site
|
|
|
| 35 | c3lerA_ |
|
not modelled |
100.0 |
30 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from campylobacter2 jejuni subsp. jejuni nctc 11168
|
|
|
| 36 | c5c54D_ |
|
not modelled |
100.0 |
24 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase/n-acetylneuraminate lyase;
PDBTitle: crystal structure of a novel n-acetylneuraminic acid lyase from2 corynebacterium glutamicum
|
|
|
| 37 | c2v9dB_ |
|
not modelled |
100.0 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
|
|
| 38 | c6arhA_ |
|
not modelled |
100.0 |
29 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: crystal structure of human nal at a resolution of 1.6 angstrom
|
|
|
| 39 | c3daqB_ |
|
not modelled |
100.0 |
34 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
|
|
| 40 | c3n2xB_ |
|
not modelled |
100.0 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
|
|
| 41 | c3e96B_ |
|
not modelled |
100.0 |
28 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bacillus2 clausii
|
|
|
| 42 | c3fkkA_ |
|
not modelled |
100.0 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
|
|
| 43 | c4dppB_ |
|
not modelled |
100.0 |
28 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase 2, chloroplastic;
PDBTitle: the structure of dihydrodipicolinate synthase 2 from arabidopsis2 thaliana
|
|
|
| 44 | c6daqA_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:phdj;
PDBTitle: phdj bound to substrate intermediate
|
|
|
| 45 | c3d0cB_ |
|
not modelled |
100.0 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
|
|
|
| 46 | c4ur7B_ |
|
not modelled |
100.0 |
28 |
PDB header:lyase
Chain: B: PDB Molecule:keto-deoxy-d-galactarate dehydratase;
PDBTitle: crystal structure of keto-deoxy-d-galactarate dehydratase2 complexed with pyruvate
|
|
|
| 47 | d1w3ia_ |
|
not modelled |
100.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 48 | c5ui3C_ |
|
not modelled |
100.0 |
28 |
PDB header:lyase
Chain: C: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dhdps from chlamydomonas reinhardtii
|
|
|
| 49 | c2r94B_ |
|
not modelled |
100.0 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
|
|
| 50 | c3dz1A_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
|
|
| 51 | c2nuxB_ |
|
not modelled |
100.0 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconate
PDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
|
|
|
| 52 | c4uxdC_ |
|
not modelled |
100.0 |
21 |
PDB header:lyase
Chain: C: PDB Molecule:2-dehydro-3-deoxy-d-gluconate/2-dehydro-3-deoxy-
PDBTitle: 2-keto 3-deoxygluconate aldolase from picrophilus torridus
|
|
|
| 53 | c2pcqA_ |
|
not modelled |
100.0 |
32 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthase;
PDBTitle: crystal structure of putative dihydrodipicolinate synthase (ttha0737)2 from thermus thermophilus hb8
|
|
|
| 54 | c6daoB_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:trans-o-hydroxybenzylidenepyruvate hydratase-aldolase;
PDBTitle: nahe wt selenomethionine
|
|
|
| 55 | c3b4uB_ |
|
not modelled |
100.0 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
|
|
| 56 | c2hmcA_ |
|
not modelled |
100.0 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
|
|
| 57 | c3qfeB_ |
|
not modelled |
100.0 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
|
|
| 58 | c4dnhA_ |
|
not modelled |
98.8 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of hypothetical protein smc04132 from sinorhizobium2 meliloti 1021
|
|
|
| 59 | d1muma_ |
|
not modelled |
98.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 60 | c2ze3A_ |
|
not modelled |
98.1 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
|
|
| 61 | c3lyeA_ |
|
not modelled |
98.1 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
|
|
| 62 | d1ujqa_ |
|
not modelled |
98.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 63 | c3ih1A_ |
|
not modelled |
97.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
|
|
| 64 | c3w9zA_ |
|
not modelled |
97.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine synthase c;
PDBTitle: crystal structure of dusc
|
|
|
| 65 | c3eooL_ |
|
not modelled |
97.8 |
16 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
|
|
| 66 | c4lsbA_ |
|
not modelled |
97.8 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:lyase/mutase;
PDBTitle: crystal structure of a putative lyase/mutase from burkholderia2 cenocepacia j2315
|
|
|
| 67 | c3fa4D_ |
|
not modelled |
97.8 |
18 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
|
|
| 68 | c4mg4G_ |
|
not modelled |
97.8 |
14 |
PDB header:unknown function
Chain: G: PDB Molecule:phosphonomutase;
PDBTitle: crystal structure of a putative phosphonomutase from burkholderia2 cenocepacia j2315
|
|
|
| 69 | c5uncB_ |
|
not modelled |
97.8 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoenolpyruvate phosphomutase;
PDBTitle: the crystal structure of phosphoenolpyruvate phosphomutase from2 streptomyces platensis subsp. rosaceus
|
|
|
| 70 | c2qiwA_ |
|
not modelled |
97.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
|
|
| 71 | d1s2wa_ |
|
not modelled |
97.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 72 | c2hjpA_ |
|
not modelled |
97.8 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
|
|
| 73 | c1zlpA_ |
|
not modelled |
97.7 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
|
|
| 74 | c3b8iF_ |
|
not modelled |
97.7 |
23 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
|
|
| 75 | d1xcfa_ |
|
not modelled |
97.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 76 | c2zviB_ |
|
not modelled |
97.5 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate
PDBTitle: crystal structure of 2,3-diketo-5-methylthiopentyl-1-2 phosphate enolase from bacillus subtilis
|
|
|
| 77 | c1rcxH_ |
|
not modelled |
97.3 |
18 |
PDB header:lyase (carbon-carbon)
Chain: H: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase;
PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
|
|
|
| 78 | c4nasD_ |
|
not modelled |
97.3 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:ribulose-bisphosphate carboxylase;
PDBTitle: the crystal structure of a rubisco-like protein (mtnw) from2 alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
|
|
|
| 79 | c5ocsB_ |
|
not modelled |
97.3 |
20 |
PDB header:flavoprotein
Chain: B: PDB Molecule:putative nadh-depentdent flavin oxidoreductase;
PDBTitle: ene-reductase (er/oye) from ralstonia (cupriavidus) metallidurans
|
|
|
| 80 | c3navB_ |
|
not modelled |
97.3 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 81 | c5n2pA_ |
|
not modelled |
97.3 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
| 82 | c3gr7A_ |
|
not modelled |
97.2 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal crystal form
|
|
|
| 83 | d1rbla1 |
|
not modelled |
97.2 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 84 | d1ykwa1 |
|
not modelled |
97.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 85 | c2h90A_ |
|
not modelled |
97.2 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xenobiotic reductase a;
PDBTitle: xenobiotic reductase a in complex with coumarin
|
|
|
| 86 | d1ej7l1 |
|
not modelled |
97.2 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 87 | c4xp7A_ |
|
not modelled |
97.1 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine(20) synthase [nad(p)+]-like;
PDBTitle: crystal structure of human trna dihydrouridine synthase 2
|
|
|
| 88 | d1svda1 |
|
not modelled |
97.1 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 89 | c3ez4B_ |
|
not modelled |
97.1 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate hydroxymethyltransferase2 from burkholderia pseudomallei
|
|
|
| 90 | d8ruca1 |
|
not modelled |
97.1 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 91 | c3fk4A_ |
|
not modelled |
97.1 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:rubisco-like protein;
PDBTitle: crystal structure of rubisco-like protein from bacillus2 cereus atcc 14579
|
|
|
| 92 | c2oemA_ |
|
not modelled |
97.0 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate enolase;
PDBTitle: crystal structure of a rubisco-like protein from geobacillus2 kaustophilus liganded with mg2+ and 2,3-diketohexane 1-phosphate
|
|
|
| 93 | d2d69a1 |
|
not modelled |
97.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 94 | c2qygC_ |
|
not modelled |
97.0 |
14 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
|
|
| 95 | c1ps9A_ |
|
not modelled |
97.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-dienoyl2 coa reductase
|
|
|
| 96 | d1z41a1 |
|
not modelled |
97.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 97 | d1oy0a_ |
|
not modelled |
97.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 98 | c6ei9A_ |
|
not modelled |
97.0 |
15 |
PDB header:flavoprotein
Chain: A: PDB Molecule:trna-dihydrouridine synthase b;
PDBTitle: crystal structure of e. coli trna-dihydrouridine synthase b (dusb)
|
|
|
| 99 | d1geha1 |
|
not modelled |
96.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 100 | d1ps9a1 |
|
not modelled |
96.9 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 101 | c5zknA_ |
|
not modelled |
96.9 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate 2-epimerase from2 fusobacterium nucleatum
|
|
|
| 102 | d1wdda1 |
|
not modelled |
96.9 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 103 | d1gk8a1 |
|
not modelled |
96.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 104 | c6hunA_ |
|
not modelled |
96.9 |
14 |
PDB header:photosynthesis
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: dimeric archeal rubisco from hyperthermus butylicus
|
|
|
| 105 | c3nwrA_ |
|
not modelled |
96.8 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:a rubisco-like protein;
PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
|
|
|
| 106 | c1gehE_ |
|
not modelled |
96.8 |
13 |
PDB header:lyase
Chain: E: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase;
PDBTitle: crystal structure of archaeal rubisco (ribulose 1,5-bisphosphate2 carboxylase/oxygenase)
|
|
|
| 107 | c3bolB_ |
|
not modelled |
96.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
|
|
| 108 | c3thaB_ |
|
not modelled |
96.7 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
|
|
|
| 109 | c4ot7A_ |
|
not modelled |
96.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of a variant of ncr from zymomonas mobilis
|
|
|
| 110 | c1bwvA_ |
|
not modelled |
96.7 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:protein (ribulose bisphosphate carboxylase);
PDBTitle: activated ribulose 1,5-bisphosphate carboxylase/oxygenase (rubisco)2 complexed with the reaction intermediate analogue 2-carboxyarabinitol3 1,5-bisphosphate
|
|
|
| 111 | c5ey5A_ |
|
not modelled |
96.7 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:lbcats-a;
PDBTitle: lbcats
|
|
|
| 112 | c2rduA_ |
|
not modelled |
96.7 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyacid oxidase 1;
PDBTitle: crystal structure of human glycolate oxidase in complex with2 glyoxylate
|
|
|
| 113 | c1rldB_ |
|
not modelled |
96.7 |
13 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:ribulose 1,5 bisphosphate carboxylase/oxygenase (large
PDBTitle: solid-state phase transition in the crystal structure of ribulose 1,5-2 biphosphate carboxylase(slash)oxygenase
|
|
|
| 114 | c3hf3A_ |
|
not modelled |
96.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
|
|
| 115 | d1qopa_ |
|
not modelled |
96.6 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 116 | c2e77B_ |
|
not modelled |
96.6 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lactate oxidase;
PDBTitle: crystal structure of l-lactate oxidase with pyruvate complex
|
|
|
| 117 | d1tv5a1 |
|
not modelled |
96.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 118 | c1tv5A_ |
|
not modelled |
96.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydroorotate dehydrogenase homolog, mitochondrial;
PDBTitle: plasmodium falciparum dihydroorotate dehydrogenase with a bound2 inhibitor
|
|
|
| 119 | c2rdtA_ |
|
not modelled |
96.6 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyacid oxidase 1;
PDBTitle: crystal structure of human glycolate oxidase (go) in complex with cdst
|
|
|
| 120 | d1f76a_ |
|
not modelled |
96.6 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|