1 c2af6G_
100.0
98
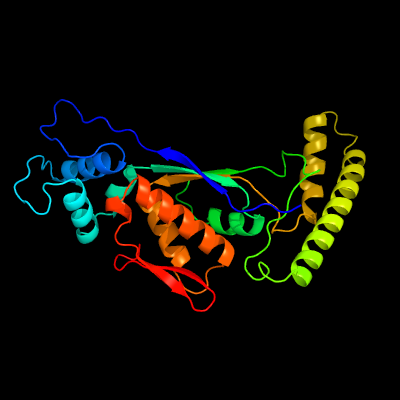
PDB header: transferaseChain: G: PDB Molecule: thymidylate synthase thyx;PDBTitle: crystal structure of mycobacterium tuberculosis flavin dependent2 thymidylate synthase (mtb thyx) in the presence of co-factor fad and3 substrate analog 5-bromo-2'-deoxyuridine-5'-monophosphate (brdump)
2 c3fnnA_
100.0
64
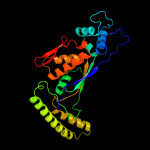
PDB header: transferaseChain: A: PDB Molecule: thymidylate synthase thyx;PDBTitle: biochemical and structural analysis of an atypical thyx:2 corynebacterium glutamicum nchu 87078 depends on thya for3 thymidine biosynthesis
3 c6j61B_
100.0
24
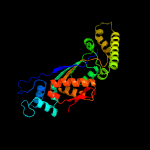
PDB header: transferaseChain: B: PDB Molecule: flavin-dependent thymidylate synthase;PDBTitle: crystal structure of thymidylate synthase, thy1, from thermus2 thermophilus having an extra c terminal domain
4 d1o26a_
100.0
21
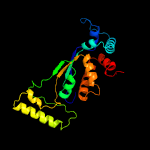
Fold: Thymidylate synthase-complementing protein Thy1Superfamily: Thymidylate synthase-complementing protein Thy1Family: Thymidylate synthase-complementing protein Thy1
5 c3ah5E_
100.0
25
PDB header: transferaseChain: E: PDB Molecule: thymidylate synthase thyx;PDBTitle: crystal structure of flavin dependent thymidylate synthase thyx from2 helicobacter pylori complexed with fad and dump
6 c4p5aB_
100.0
23
PDB header: transferaseChain: B: PDB Molecule: thymidylate synthase thyx;PDBTitle: crystal structure of a ump/dump methylase polb from streptomyces2 cacaoi bound with 5-br ump
7 c2cfaB_
100.0
31
PDB header: transferaseChain: B: PDB Molecule: thymidylate synthase;PDBTitle: structure of viral flavin-dependant thymidylate synthase thyx
8 c3cuxA_
25.2
35
PDB header: transferaseChain: A: PDB Molecule: malate synthase;PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
9 c5ydfA_
19.1
25
PDB header: transcriptionChain: A: PDB Molecule: parafibromin;PDBTitle: crystal structure of a disease-related gene, hcdc73(1-100)
10 c3cuzA_
14.9
38
PDB header: transferaseChain: A: PDB Molecule: malate synthase a;PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
11 d2v7fa1
14.0
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rps19E-like
12 c3mmiB_
12.8
11
PDB header: motor proteinChain: B: PDB Molecule: myosin-4;PDBTitle: crystal structure of the globular tail of myo4p
13 c5vfbB_
12.0
31
PDB header: transferaseChain: B: PDB Molecule: malate synthase g;PDBTitle: 1.36 angstrom resolution crystal structure of malate synthase g from2 pseudomonas aeruginosa in complex with glycolic acid.
14 d1nb5i_
11.2
21
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cystatins
15 c4i1oD_
10.5
16
PDB header: protein transport/hydrolaseChain: D: PDB Molecule: lepb;PDBTitle: crystal structure of the legionella pneumophila gap domain of lepb in2 complex with rab1b bound to gdp and bef3
16 c3j21a_
10.5
9
PDB header: ribosomeChain: A: PDB Molecule: 50s ribosomal protein l1p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
17 c3mlqD_
9.8
35
PDB header: transferase/transcriptionChain: D: PDB Molecule: dna-directed rna polymerase subunit beta;PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
18 c5tpjA_
9.5
32
PDB header: de novo proteinChain: A: PDB Molecule: denovo ntf2;PDBTitle: crystal structure of a de novo designed protein with curved beta-sheet
19 c5gyqA_
9.2
17
PDB header: viral proteinChain: A: PDB Molecule: spike glycoprotein;PDBTitle: putative receptor-binding domain of bat-derived coronavirus hku9 spike2 protein
20 d2np3a1
8.8
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain
21 d1vqox1
not modelled
8.8
25
Fold: Ribosomal protein L31eSuperfamily: Ribosomal protein L31eFamily: Ribosomal protein L31e
22 d1d1da1
not modelled
7.5
19
Fold: Acyl carrier protein-likeSuperfamily: Retrovirus capsid dimerization domain-likeFamily: Retrovirus capsid protein C-terminal domain
23 c5lphA_
not modelled
7.4
19
PDB header: cell cycleChain: A: PDB Molecule: centrosomal protein of 104 kda;PDBTitle: structure of the tog domain of human cep104
24 c2xalA_
not modelled
7.4
30
PDB header: transferaseChain: A: PDB Molecule: inositol-pentakisphosphate 2-kinase;PDBTitle: lead derivative of inositol 1,3,4,5,6-pentakisphosphate 2-kinase from2 a. thaliana in complex with adp and ip6.
25 c5vbnE_
not modelled
7.4
21
PDB header: transferaseChain: E: PDB Molecule: dna polymerase epsilon subunit 2;PDBTitle: crystal structure of human dna polymerase epsilon b-subunit in complex2 with c-terminal domain of catalytic subunit
26 c2n28A_
not modelled
7.3
11
PDB header: viral proteinChain: A: PDB Molecule: protein vpu;PDBTitle: solid-state nmr structure of vpu
27 c3pp5A_
not modelled
7.0
11
PDB header: structural proteinChain: A: PDB Molecule: brk1;PDBTitle: high-resolution structure of the trimeric scar/wave complex precursor2 brk1
28 c1yylS_
not modelled
7.0
30
PDB header: viral protein/immune systemChain: S: PDB Molecule: cd4m33, scorpion-toxin mimic of cd4;PDBTitle: crystal structure of cd4m33, a scorpion-toxin mimic of cd4, in complex2 with hiv-1 yu2 gp120 envelope glycoprotein and anti-hiv-1 antibody3 17b
29 c1yylM_
not modelled
7.0
30
PDB header: viral protein/immune systemChain: M: PDB Molecule: cd4m33, scorpion-toxin mimic of cd4;PDBTitle: crystal structure of cd4m33, a scorpion-toxin mimic of cd4, in complex2 with hiv-1 yu2 gp120 envelope glycoprotein and anti-hiv-1 antibody3 17b
30 c4r4ns_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: S: PDB Molecule: hiv-1 gp120;PDBTitle: crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-2 1 93ug037 gp120
31 c4r4ni_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: I: PDB Molecule: hiv-1 gp120;PDBTitle: crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-2 1 93ug037 gp120
32 c5w4lM_
not modelled
6.9
30
PDB header: viral protein/immune systemChain: M: PDB Molecule: cd4 mimetic peptide m48u1;PDBTitle: crystal structure of the non-neutralizing and adcc-potent c11-like2 antibody n12-i3 in complex with hiv-1 clade a/e gp120, the cd43 mimetic m48u1, and the antibody n5-i5.
33 c4ka2R_
not modelled
6.9
30
PDB header: viral protein/inhibitorChain: R: PDB Molecule: m48u12;PDBTitle: crystal structure of cd4-mimetic miniprotein m48u12 in complex with2 hiv-1 yu2 gp120
34 c4r4na_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: A: PDB Molecule: hiv-1 gp120;PDBTitle: crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-2 1 93ug037 gp120
35 c4r4nb_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: B: PDB Molecule: hiv-1 gp120;PDBTitle: crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-2 1 93ug037 gp120
36 c5kjrN_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: N: PDB Molecule: m48u1 cd4 mimetic peptide;PDBTitle: crystal structure of the adcc-potent antibody n60-i3 fab in complex2 with hiv-1 clade a/e gp120 w69a/s115w mutant and m48u1.
37 c4k0aR_
not modelled
6.9
30
PDB header: viral protein/inhibitorChain: R: PDB Molecule: cd4-mimetic miniprotein m48u7;PDBTitle: crystal structure of cd4-mimetic miniprotein m48u7 in complex with2 hiv-1 yu2 gp120
38 c4r4nm_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: M: PDB Molecule: hiv-1 gp120;PDBTitle: crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-2 1 93ug037 gp120
39 c4jzzR_
not modelled
6.9
30
PDB header: viral protein/inhibitorChain: R: PDB Molecule: cd4-mimetic miniprotein m48u1;PDBTitle: crystal structure of cd4-mimetic miniprotein m48u1 in complex with2 hiv-1 yu2 gp120 in c2221 space group
40 c4r4nv_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: V: PDB Molecule: hiv-1 gp120;PDBTitle: crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-2 1 93ug037 gp120
41 c4rfoN_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: N: PDB Molecule: m48u1 cd4 mimetic peptide;PDBTitle: crystal structure of the adcc-potent antibody n60-i3 fab in complex2 with hiv-1 clade a/e gp120 and m48u1
42 c4jzwR_
not modelled
6.9
30
PDB header: viral protein/inhibitorChain: R: PDB Molecule: cd4-mimetic miniprotein m48u1;PDBTitle: crystal structure of cd4-mimetic miniprotein m48u1 in complex with2 hiv-1 yu2 gp120 in p212121 space group
43 c4lajK_
not modelled
6.9
30
PDB header: viral protein/inhibitorChain: K: PDB Molecule: cd4-mimetic miniprotein m48u1;PDBTitle: crystal structure of hiv-1 yu2 envelope gp120 glycoprotein in complex2 with cd4-mimetic miniprotein, m48u1, and llama single-domain, broadly3 neutralizing, co-receptor binding site antibody, jm4
44 c5uweN_
not modelled
6.9
30
PDB header: viral protein/immune systemChain: N: PDB Molecule: m48u1 cd4 mimetic peptide;PDBTitle: crystal structure of the adcc-potent, weakly neutralizing hiv env co-2 receptor binding site antibody n12-i2 fab in complex with hiv-1 clade3 a/e gp120 and m48u1
45 c4r4np_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: P: PDB Molecule: hiv-1 gp120;PDBTitle: crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-2 1 93ug037 gp120
46 c4r4ne_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: E: PDB Molecule: hiv-1 gp120;PDBTitle: crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-2 1 93ug037 gp120
47 c4r4fR_
not modelled
6.9
30
PDB header: viral protein/immune system/inhibitorChain: R: PDB Molecule: m48u1 peptide;PDBTitle: crystal structure of non-neutralizing, a32-like antibody 2.2c in2 complex with hiv-1 yu2 gp120
48 c4lajC_
not modelled
6.9
30
PDB header: viral protein/inhibitorChain: C: PDB Molecule: cd4-mimetic miniprotein m48u1;PDBTitle: crystal structure of hiv-1 yu2 envelope gp120 glycoprotein in complex2 with cd4-mimetic miniprotein, m48u1, and llama single-domain, broadly3 neutralizing, co-receptor binding site antibody, jm4
49 c5w4lN_
not modelled
6.9
30
PDB header: viral protein/immune systemChain: N: PDB Molecule: cd4 mimetic peptide m48u1;PDBTitle: crystal structure of the non-neutralizing and adcc-potent c11-like2 antibody n12-i3 in complex with hiv-1 clade a/e gp120, the cd43 mimetic m48u1, and the antibody n5-i5.
50 c4lajG_
not modelled
6.9
30
PDB header: viral protein/inhibitorChain: G: PDB Molecule: cd4-mimetic miniprotein m48u1;PDBTitle: crystal structure of hiv-1 yu2 envelope gp120 glycoprotein in complex2 with cd4-mimetic miniprotein, m48u1, and llama single-domain, broadly3 neutralizing, co-receptor binding site antibody, jm4
51 c2i5yM_
not modelled
6.8
30
PDB header: viral protein/immune systemChain: M: PDB Molecule: cd4m47, scorpion-toxin mimic of cd4;PDBTitle: crystal structure of cd4m47, a scorpion-toxin mimic of cd4, in complex2 with hiv-1 yu2 gp120 envelope glycoprotein and anti-hiv-1 antibody3 17b
52 c2p9lD_
not modelled
6.8
19
PDB header: structural proteinChain: D: PDB Molecule: actin-related protein 2/3 complex subunit 2;PDBTitle: crystal structure of bovine arp2/3 complex
53 c3bbnD_
not modelled
6.7
21
PDB header: ribosomeChain: D: PDB Molecule: ribosomal protein s4;PDBTitle: homology model for the spinach chloroplast 30s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome.
54 c1yymS_
not modelled
6.7
30
PDB header: viral protein/immune systemChain: S: PDB Molecule: f23, scorpion-toxin mimic of cd4;PDBTitle: crystal structure of f23, a scorpion-toxin mimic of cd4, in complex2 with hiv-1 yu2 gp120 envelope glycoprotein and anti-hiv-1 antibody3 17b
55 c1yymM_
not modelled
6.7
30
PDB header: viral protein/immune systemChain: M: PDB Molecule: f23, scorpion-toxin mimic of cd4;PDBTitle: crystal structure of f23, a scorpion-toxin mimic of cd4, in complex2 with hiv-1 yu2 gp120 envelope glycoprotein and anti-hiv-1 antibody3 17b
56 c4rfnD_
not modelled
6.7
30
PDB header: viral protein/immune systemChain: D: PDB Molecule: t-cell surface glycoprotein cd4 mimetic m48;PDBTitle: crystal structure of adcc-potent rhesus macaque antibody jr4 in2 complex with hiv-1 clade a/e gp120 and m48
57 c4rfnM_
not modelled
6.7
30
PDB header: viral protein/immune systemChain: M: PDB Molecule: t-cell surface glycoprotein cd4 mimetic m48;PDBTitle: crystal structure of adcc-potent rhesus macaque antibody jr4 in2 complex with hiv-1 clade a/e gp120 and m48
58 c2i60M_
not modelled
6.6
30
PDB header: viral protein/immune systemChain: M: PDB Molecule: [phe23]m47, scorpion-toxin mimic of cd4;PDBTitle: crystal structure of [phe23]m47, a scorpion-toxin mimic of cd4, in2 complex with hiv-1 yu2 gp120 envelope glycoprotein and anti-hiv-13 antibody 17b
59 c2i60S_
not modelled
6.6
30
PDB header: viral protein/immune systemChain: S: PDB Molecule: [phe23]m47, scorpion-toxin mimic of cd4;PDBTitle: crystal structure of [phe23]m47, a scorpion-toxin mimic of cd4, in2 complex with hiv-1 yu2 gp120 envelope glycoprotein and anti-hiv-13 antibody 17b
60 d1n8ia_
not modelled
6.4
33
Fold: TIM beta/alpha-barrelSuperfamily: Malate synthase GFamily: Malate synthase G
61 c3udsA_
not modelled
6.1
30
PDB header: transferaseChain: A: PDB Molecule: inositol-pentakisphosphate 2-kinase;PDBTitle: inositol 1,3,4,5,6-pentakisphosphate 2-kinase from a. thaliana in2 complex with adp.
62 c3pf6C_
not modelled
5.6
38
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein pp-luz7_gp033;PDBTitle: the structure of uncharacterized protein pp-luz7_gp033 from2 pseudomonas phage luz7.
63 d1k8kd2
not modelled
5.2
19
Fold: Secretion chaperone-likeSuperfamily: Arp2/3 complex subunitsFamily: Arp2/3 complex subunits
64 c3izrg_
not modelled
5.2
20
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein l6 (l6e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome