| 1 |
|
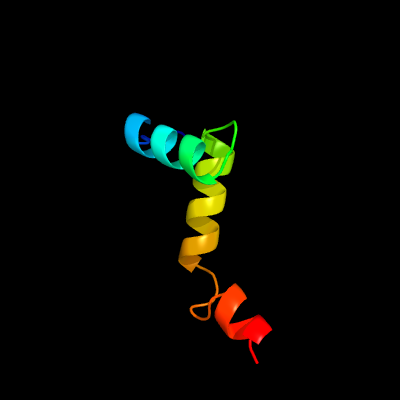
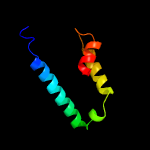
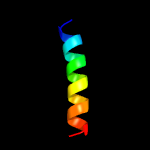
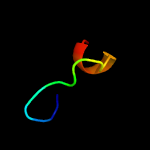
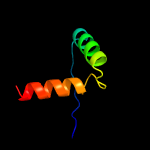
PDB 6a7v chain U
Region: 16 - 62
Aligned: 47
Modelled: 47
Confidence: 98.0%
Identity: 38%
PDB header:toxin/antitoxin
Chain: U: PDB Molecule:antitoxin vapb11;
PDBTitle: crystal structure of mycobacterium tuberculosis vapbc11 toxin-2 antitoxin complex
Phyre2
| 2 |
|
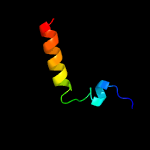
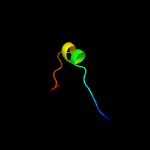
PDB 1ib8 chain A
Region: 1 - 23
Aligned: 23
Modelled: 23
Confidence: 37.7%
Identity: 30%
PDB header:nucleic acid binding protein
Chain: A: PDB Molecule:conserved protein sp14.3;
PDBTitle: solution structure and function of a conserved protein2 sp14.3 encoded by an essential streptococcus pneumoniae3 gene
Phyre2
| 3 |
|
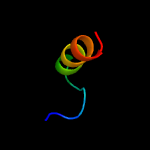
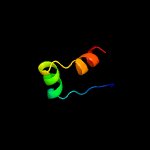
PDB 3nct chain C
Region: 2 - 13
Aligned: 12
Modelled: 12
Confidence: 24.3%
Identity: 50%
PDB header:dna binding protein, chaperone
Chain: C: PDB Molecule:protein psib;
PDBTitle: x-ray crystal structure of the bacterial conjugation factor psib, a2 negative regulator of reca
Phyre2
| 4 |
|
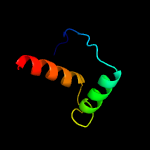
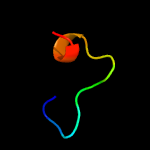
PDB 3nvt chain A
Region: 16 - 81
Aligned: 62
Modelled: 66
Confidence: 23.0%
Identity: 19%
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;
PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
Phyre2
| 5 |
|
PDB 5gl6 chain A
Region: 5 - 23
Aligned: 19
Modelled: 19
Confidence: 19.3%
Identity: 42%
PDB header:ribosomal protein
Chain: A: PDB Molecule:ribosome maturation factor rimp;
PDBTitle: msmeg rimp
Phyre2
| 6 |
|
PDB 4e6f chain B
Region: 15 - 34
Aligned: 20
Modelled: 20
Confidence: 16.7%
Identity: 30%
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a duf4468 family protein (bacova_04320) from2 bacteroides ovatus atcc 8483 at 1.49 a resolution
Phyre2
| 7 |
|
PDB 3f5h chain B
Region: 23 - 53
Aligned: 31
Modelled: 31
Confidence: 14.2%
Identity: 42%
PDB header:protein binding
Chain: B: PDB Molecule:type i polyketide synthase pikaiii, type i polyketide
PDBTitle: crystal structure of fused docking domains from pikaiii and pikaiv of2 the pikromycin polyketide synthase
Phyre2
| 8 |
|
PDB 3lj4 chain I
Region: 37 - 55
Aligned: 19
Modelled: 19
Confidence: 13.8%
Identity: 58%
PDB header:viral protein
Chain: I: PDB Molecule:portal protein;
PDBTitle: bacteriophage p22 portal protein bound to middle tail factor gp4. this2 file contain the first biological assembly
Phyre2
| 9 |
|
PDB 2k5j chain B
Region: 10 - 56
Aligned: 45
Modelled: 47
Confidence: 10.7%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein yiif;
PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
Phyre2
| 10 |
|
PDB 2eho chain L
Region: 11 - 25
Aligned: 15
Modelled: 15
Confidence: 10.5%
Identity: 27%
PDB header:replication
Chain: L: PDB Molecule:gins complex subunit 3;
PDBTitle: crystal structure of human gins complex
Phyre2
| 11 |
|
PDB 2qs7 chain D
Region: 4 - 23
Aligned: 20
Modelled: 20
Confidence: 9.9%
Identity: 25%
PDB header:oxidoreductase
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative oxidoreductase of the dsre/dsrf-like2 family (sso1126) from sulfolobus solfataricus p2 at 2.09 a resolution
Phyre2
| 12 |
|
PDB 2jfg chain A domain 1
Region: 4 - 32
Aligned: 24
Modelled: 29
Confidence: 9.0%
Identity: 38%
Fold: MurCD N-terminal domain
Superfamily: MurCD N-terminal domain
Family: MurCD N-terminal domain
Phyre2
| 13 |
|
PDB 5gqs chain A
Region: 6 - 14
Aligned: 9
Modelled: 9
Confidence: 7.9%
Identity: 44%
PDB header:transport protein
Chain: A: PDB Molecule:pts galactitol transporter subunit iib;
PDBTitle: nmr based solution structure of pts system, galactitol-specific iib2 component from methicillin resistant staphylococcus aureus
Phyre2
| 14 |
|
PDB 2e9x chain C domain 2
Region: 11 - 25
Aligned: 15
Modelled: 15
Confidence: 7.8%
Identity: 27%
Fold: GINS/PriA/YqbF domain
Superfamily: PriA/YqbF domain
Family: PSF3 N-terminal domain-like
Phyre2
| 15 |
|
PDB 2l2q chain A
Region: 7 - 19
Aligned: 13
Modelled: 13
Confidence: 7.2%
Identity: 38%
PDB header:transferase
Chain: A: PDB Molecule:pts system, cellobiose-specific iib component (cela);
PDBTitle: solution structure of cellobiose-specific phosphotransferase iib2 component protein from borrelia burgdorferi
Phyre2
| 16 |
|
PDB 6ajn chain F
Region: 22 - 59
Aligned: 38
Modelled: 38
Confidence: 6.8%
Identity: 13%
PDB header:toxin
Chain: F: PDB Molecule:duf1778 domain-containing protein;
PDBTitle: crystal structure of atatr bound with accoa
Phyre2
| 17 |
|
PDB 2bj7 chain A domain 1
Region: 12 - 56
Aligned: 44
Modelled: 45
Confidence: 6.5%
Identity: 20%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: CopG-like
Phyre2
| 18 |
|
PDB 6av8 chain A
Region: 8 - 12
Aligned: 5
Modelled: 5
Confidence: 6.2%
Identity: 80%
PDB header:toxin
Chain: A: PDB Molecule:u5-theraphotoxin-hs1b 1;
PDBTitle: exploring cystine dense peptide space to open a unique molecular2 toolbox
Phyre2
| 19 |
|
PDB 1zbs chain A domain 2
Region: 17 - 34
Aligned: 18
Modelled: 18
Confidence: 6.0%
Identity: 28%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: BadF/BadG/BcrA/BcrD-like
Phyre2
| 20 |
|
PDB 4nux chain A
Region: 2 - 13
Aligned: 12
Modelled: 12
Confidence: 5.8%
Identity: 33%
PDB header:immune system
Chain: A: PDB Molecule:interleukin-17 receptor a;
PDBTitle: structure of receptor a
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|