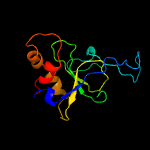
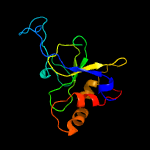
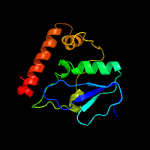
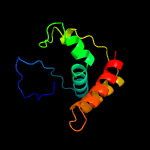
1 c1yf2A_
100.0
16
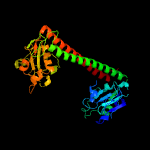
PDB header: hydrolase regulatorChain: A: PDB Molecule: type i restriction-modification enzyme, s subunit;PDBTitle: three-dimensional structure of dna sequence specificity (s) subunit of2 a type i restriction-modification enzyme and its functional3 implications
2 c1ydxA_
100.0
16
PDB header: dna binding proteinChain: A: PDB Molecule: type i restriction enzyme specificity protein mg438;PDBTitle: crystal structure of type-i restriction-modification system s subunit2 from m. genitalium
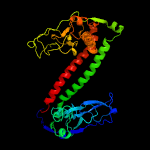
3 c2y7cA_
100.0
10
PDB header: transferaseChain: A: PDB Molecule: type-1 restriction enzyme ecoki specificity protein;PDBTitle: atomic model of the ocr-bound methylase complex from the type i2 restriction-modification enzyme ecoki (m2s1). based on fitting into3 em map 1534.
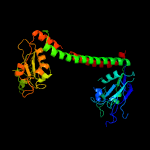
4 c3okgB_
100.0
12
PDB header: dna binding proteinChain: B: PDB Molecule: restriction endonuclease s subunits;PDBTitle: crystal structure of hsds subunit from thermoanaerobacter2 tengcongensis
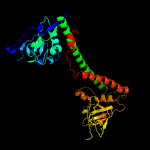
5 d1ydxa2
100.0
18
Fold: DNA methylase specificity domainSuperfamily: DNA methylase specificity domainFamily: Type I restriction modification DNA specificity domain
6 d1yf2a2
99.9
16
Fold: DNA methylase specificity domainSuperfamily: DNA methylase specificity domainFamily: Type I restriction modification DNA specificity domain
7 d1ydxa1
99.9
19
Fold: DNA methylase specificity domainSuperfamily: DNA methylase specificity domainFamily: Type I restriction modification DNA specificity domain
8 d1yf2a1
99.9
17
Fold: DNA methylase specificity domainSuperfamily: DNA methylase specificity domainFamily: Type I restriction modification DNA specificity domain
9 c1aqjB_
97.1
13
PDB header: methyltransferaseChain: B: PDB Molecule: adenine-n6-dna-methyltransferase taqi;PDBTitle: structure of adenine-n6-dna-methyltransferase taqi
10 c1g38A_
96.8
13
PDB header: transferase/dnaChain: A: PDB Molecule: modification methylase taqi;PDBTitle: adenine-specific methyltransferase m. taq i/dna complex
11 c3s1sA_
93.3
13
PDB header: hydrolase, transferaseChain: A: PDB Molecule: restriction endonuclease bpusi;PDBTitle: characterization and crystal structure of the type iig restriction2 endonuclease bpusi
12 c5hr4J_
76.3
13
PDB header: hydrolase/dnaChain: J: PDB Molecule: mmei;PDBTitle: structure of type iil restriction-modification enzyme mmei in complex2 with dna has implications for engineering of new specificities
13 c3bc1F_
49.9
12
PDB header: signaling protein/transport proteinChain: F: PDB Molecule: synaptotagmin-like protein 2;PDBTitle: crystal structure of the complex rab27a-slp2a
14 d1zkea1
41.9
7
Fold: ROP-likeSuperfamily: HP1531-likeFamily: HP1531-like
15 c5ihfA_
32.3
20
PDB header: unknown functionChain: A: PDB Molecule: virg-like protein;PDBTitle: salmonella typhimurium virg-like (stv) protein
16 c4jkvA_
26.7
20
PDB header: membrane proteinChain: A: PDB Molecule: soluble cytochrome b562, smoothened homolog;PDBTitle: structure of the human smoothened 7tm receptor in complex with an2 antitumor agent
17 c4ug1A_
25.1
11
PDB header: cell cycleChain: A: PDB Molecule: cell cycle protein gpsb;PDBTitle: gpsb n-terminal domain
18 c6d80A_
23.6
6
PDB header: transport proteinChain: A: PDB Molecule: mitochondrial calcium uniporter;PDBTitle: cryo-em structure of the mitochondrial calcium uniporter from n.2 fischeri bound to saposin
19 c2lqtA_
22.5
22
PDB header: unknown functionChain: A: PDB Molecule: coiled-coil-helix-coiled-coil-helix domain-containingPDBTitle: solution structure of chchd7
20 d2ih2a2
21.5
14
Fold: DNA methylase specificity domainSuperfamily: DNA methylase specificity domainFamily: TaqI C-terminal domain-like
21 c6j5id_
not modelled
16.8
18
PDB header: membrane proteinChain: D: PDB Molecule: atp synthase subunit beta;PDBTitle: cryo-em structure of the mammalian dp-state atp synthase
22 c6roiA_
not modelled
16.4
18
PDB header: lipid transportChain: A: PDB Molecule: probable phospholipid-transporting atpase drs2;PDBTitle: cryo-em structure of the partially activated drs2p-cdc50p
23 c3vhlA_
not modelled
16.2
16
PDB header: signaling proteinChain: A: PDB Molecule: dedicator of cytokinesis protein 8;PDBTitle: crystal structure of the dhr-2 domain of dock8 in complex with cdc422 (t17n mutant)
24 c4zrkH_
not modelled
16.1
42
PDB header: signaling protein/transferaseChain: H: PDB Molecule: serine/threonine-protein kinase lats1;PDBTitle: merlin-ferm and lats1 complex
25 c4zrkF_
not modelled
16.1
42
PDB header: signaling protein/transferaseChain: F: PDB Molecule: serine/threonine-protein kinase lats1;PDBTitle: merlin-ferm and lats1 complex
26 c4zrkE_
not modelled
16.1
42
PDB header: signaling protein/transferaseChain: E: PDB Molecule: serine/threonine-protein kinase lats1;PDBTitle: merlin-ferm and lats1 complex
27 c4zrkG_
not modelled
15.9
42
PDB header: signaling protein/transferaseChain: G: PDB Molecule: serine/threonine-protein kinase lats1;PDBTitle: merlin-ferm and lats1 complex
28 c2ke4A_
not modelled
15.3
12
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
29 c5frgA_
not modelled
15.2
7
PDB header: protein bindingChain: A: PDB Molecule: formin-binding protein 1-like;PDBTitle: the nmr structure of the cdc42-interacting region of toca1
30 d1y71a1
not modelled
14.4
15
Fold: SH3-like barrelSuperfamily: Kinase-associated protein B-likeFamily: Kinase-associated protein B-like
31 c2jufA_
not modelled
14.4
16
PDB header: gene regulationChain: A: PDB Molecule: p53-associated parkin-like cytoplasmic protein;PDBTitle: nmr solution structure of parc cph domain. nesg target2 hr3443b/sgc-toronto
32 c4f8cC_
not modelled
14.4
9
PDB header: cell cycle/protein bindingChain: C: PDB Molecule: cycle inhibiting factor;PDBTitle: structure of the cif:nedd8 complex - yersinia pseudotuberculosis cycle2 inhibiting factor in complex with human nedd8
33 c2gd3A_
not modelled
13.1
42
PDB header: unknown functionChain: A: PDB Molecule: humanin;PDBTitle: nmr structure of s14g-humanin in 30% tfe solution
34 c2wukD_
not modelled
12.6
9
PDB header: cell cycleChain: D: PDB Molecule: septum site-determining protein diviva;PDBTitle: diviva n-terminal domain, f17a mutant
35 c6cfzC_
not modelled
12.6
35
PDB header: nuclear proteinChain: C: PDB Molecule: dad2;PDBTitle: structure of the dash/dam1 complex shows its role at the yeast2 kinetochore-microtubule interface
36 c3mudA_
not modelled
12.3
14
PDB header: contractile proteinChain: A: PDB Molecule: dna repair protein xrcc4,tropomyosin alpha-1 chain;PDBTitle: structure of the tropomyosin overlap complex from chicken smooth2 muscle
37 c5zx8A_
not modelled
12.0
21
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of peptidyl-trna hydrolase from thermus thermophilus
38 c3ghgK_
not modelled
11.9
8
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
39 d2jnga1
not modelled
11.9
16
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: CPH domain
40 c5xauC_
not modelled
11.5
19
PDB header: cell adhesionChain: C: PDB Molecule: laminin subunit gamma-1;PDBTitle: crystal structure of integrin binding fragment of laminin-511
41 c2xvsA_
not modelled
10.9
8
PDB header: antitumor proteinChain: A: PDB Molecule: tetratricopeptide repeat protein 5;PDBTitle: crystal structure of human ttc5 (strap) c-terminal ob2 domain
42 c2wmoA_
not modelled
10.9
24
PDB header: cell cycleChain: A: PDB Molecule: dedicator of cytokinesis protein 9;PDBTitle: structure of the complex between dock9 and cdc42.
43 c2owyB_
not modelled
10.7
13
PDB header: dna binding proteinChain: B: PDB Molecule: recombination-associated protein rdgc;PDBTitle: the recombination-associated protein rdgc adopts a novel toroidal2 architecture for dna binding
44 c2hjqA_
not modelled
10.6
10
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein yqbf;PDBTitle: nmr structure of bacillus subtilis protein yqbf, northeast2 structural genomics target sr449
45 d2nrka1
not modelled
10.4
8
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: GrpB-like
46 c4ylyB_
not modelled
10.2
14
PDB header: hydrolaseChain: B: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of peptidyl-trna hydrolase from a gram-positive2 bacterium, staphylococcus aureus at 2.25 angstrom resolution
47 c6gqaD_
not modelled
10.0
6
PDB header: cell cycleChain: D: PDB Molecule: cell cycle protein gpsb;PDBTitle: cell division regulator s. pneumoniae gpsb
48 c3kxeD_
not modelled
9.2
21
PDB header: protein bindingChain: D: PDB Molecule: antitoxin protein pard-1;PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
49 c2l5gB_
not modelled
8.7
10
PDB header: transcription regulatorChain: B: PDB Molecule: putative uncharacterized protein ncor2;PDBTitle: co-ordinates and 1h, 13c and 15n chemical shift assignments for the2 complex of gps2 53-90 and smrt 167-207
50 c3plwA_
not modelled
8.5
33
PDB header: hydrolaseChain: A: PDB Molecule: recombination enhancement function protein;PDBTitle: ref protein from p1 bacteriophage
51 c5td8B_
not modelled
8.3
12
PDB header: replicationChain: B: PDB Molecule: kinetochore protein nuf2;PDBTitle: crystal structure of an extended dwarf ndc80 complex
52 c3zifP_
not modelled
7.5
10
PDB header: virusChain: P: PDB Molecule: pix;PDBTitle: cryo-em structures of two intermediates provide insight into2 adenovirus assembly and disassembly
53 c4ct4C_
not modelled
7.3
7
PDB header: rna binding proteinChain: C: PDB Molecule: ccr4-not transcription complex subunit 1;PDBTitle: cnot1 mif4g domain - ddx6 complex
54 c4q55B_
not modelled
7.3
21
PDB header: hydrolaseChain: B: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of peptidyl-trna hydrolase from a gram-positive2 bacterium, streptococcus pyogenes at 2.19a resolution shows the3 closed structure of the substrate binding cleft
55 c3q4fG_
not modelled
7.3
17
PDB header: dna binding protein/protein bindingChain: G: PDB Molecule: dna repair protein xrcc4;PDBTitle: crystal structure of xrcc4/xlf-cernunnos complex
56 c4hpqE_
not modelled
7.3
22
PDB header: protein transportChain: E: PDB Molecule: atg31;PDBTitle: crystal structure of the atg17-atg31-atg29 complex
57 c2lvgA_
not modelled
7.2
12
PDB header: membrane proteinChain: A: PDB Molecule: non-structural protein 4b;PDBTitle: nmr structure of hcv non-structural protein ab, ns4b(1-40)
58 c3cqxD_
not modelled
6.8
24
PDB header: chaperoneChain: D: PDB Molecule: bag family molecular chaperone regulator 2;PDBTitle: chaperone complex
59 c5kb0A_
not modelled
6.8
19
PDB header: de novo proteinChain: A: PDB Molecule: pb(ii)zn(ii)(grand coil ser-l16cl30h)3+;PDBTitle: crystal structure of a tris-thiolate pb(ii) complex in a de novo2 three-stranded coiled coil peptide
60 d1q08a_
not modelled
6.8
14
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA-binding N-terminal domain of transcription activators
61 d1vf6a_
not modelled
6.7
10
Fold: L27 domainSuperfamily: L27 domainFamily: L27 domain
62 c4pn8A_
not modelled
6.7
14
PDB header: de novo proteinChain: A: PDB Molecule: cc-pent;PDBTitle: a de novo designed pentameric coiled coil cc-pent.
63 c4pn8H_
not modelled
6.7
14
PDB header: de novo proteinChain: H: PDB Molecule: cc-pent;PDBTitle: a de novo designed pentameric coiled coil cc-pent.
64 c4pn8D_
not modelled
6.7
14
PDB header: de novo proteinChain: D: PDB Molecule: cc-pent;PDBTitle: a de novo designed pentameric coiled coil cc-pent.
65 c4pn8C_
not modelled
6.7
14
PDB header: de novo proteinChain: C: PDB Molecule: cc-pent;PDBTitle: a de novo designed pentameric coiled coil cc-pent.
66 c4pn8B_
not modelled
6.7
14
PDB header: de novo proteinChain: B: PDB Molecule: cc-pent;PDBTitle: a de novo designed pentameric coiled coil cc-pent.
67 c4pn8J_
not modelled
6.7
14
PDB header: de novo proteinChain: J: PDB Molecule: cc-pent;PDBTitle: a de novo designed pentameric coiled coil cc-pent.
68 c4pn8E_
not modelled
6.7
14
PDB header: de novo proteinChain: E: PDB Molecule: cc-pent;PDBTitle: a de novo designed pentameric coiled coil cc-pent.
69 c4pn8G_
not modelled
6.7
14
PDB header: de novo proteinChain: G: PDB Molecule: cc-pent;PDBTitle: a de novo designed pentameric coiled coil cc-pent.
70 c4pn8F_
not modelled
6.7
14
PDB header: de novo proteinChain: F: PDB Molecule: cc-pent;PDBTitle: a de novo designed pentameric coiled coil cc-pent.
71 d2clyb1
not modelled
6.6
6
Fold: ATP synthase D chain-likeSuperfamily: ATP synthase D chain-likeFamily: ATP synthase D chain-like
72 c1vf6B_
not modelled
6.4
10
PDB header: protein binding/protein transportChain: B: PDB Molecule: pals1-associated tight junction protein;PDBTitle: 2.1 angstrom crystal structure of the pals-1-l27n and patj l272 heterodimer complex
73 c4melB_
not modelled
6.3
17
PDB header: hydrolaseChain: B: PDB Molecule: ubiquitin carboxyl-terminal hydrolase 11;PDBTitle: crystal structure of the human usp11 dusp-ubl domains
74 d1o5ha_
not modelled
6.3
15
Fold: Methenyltetrahydrofolate cyclohydrolase-likeSuperfamily: Methenyltetrahydrofolate cyclohydrolase-likeFamily: Methenyltetrahydrofolate cyclohydrolase-like
75 c3v2iA_
not modelled
5.8
16
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: structure of a peptidyl-trna hydrolase (pth) from burkholderia2 thailandensis
76 c1ei3E_
not modelled
5.8
9
PDB header: blood clottingChain: E: PDB Molecule: fibrinogen;PDBTitle: crystal structure of native chicken fibrinogen
77 c5gmkt_
not modelled
5.6
13
PDB header: rna binding protein/rnaChain: T: PDB Molecule: pre-mrna-splicing factor bud31;PDBTitle: cryo-em structure of the catalytic step i spliceosome (c complex) at2 3.4 angstrom resolution
78 c5gnaB_
not modelled
5.6
17
PDB header: gene regulationChain: B: PDB Molecule: flagellar hook-associated protein 2;PDBTitle: crystal structure of flagellin assembly related protein
79 c2e6iA_
not modelled
5.5
20
PDB header: transferaseChain: A: PDB Molecule: tyrosine-protein kinase itk/tsk;PDBTitle: solution structure of the btk motif of tyrosine-protein2 kinase itk from human
80 c3pp5A_
not modelled
5.5
16
PDB header: structural proteinChain: A: PDB Molecule: brk1;PDBTitle: high-resolution structure of the trimeric scar/wave complex precursor2 brk1
81 c2l2lA_
not modelled
5.5
12
PDB header: transferaseChain: A: PDB Molecule: transcriptional repressor p66-alpha;PDBTitle: solution structure of the coiled-coil complex between mbd2 and2 p66alpha
82 c3njcA_
not modelled
5.5
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: yslb protein;PDBTitle: crystal structure of the yslb protein from bacillus subtilis.2 northeast structural genomics consortium target sr460.
83 c2yinB_
not modelled
5.3
19
PDB header: apoptosisChain: B: PDB Molecule: dedicator of cytokinesis protein 2;PDBTitle: structure of the complex between dock2 and rac1.
84 c2clyE_
not modelled
5.3
6
PDB header: hydrolaseChain: E: PDB Molecule: atp synthase d chain, mitochondrial;PDBTitle: subcomplex of the stator of bovine mitochondrial atp synthase
85 c6dnfA_
not modelled
5.3
6
PDB header: membrane proteinChain: A: PDB Molecule: mitochondrial calcium uniporter mcu;PDBTitle: cryo-em structure of the mitochondrial calcium uniporter mcu from the2 fungus cyphellophora europaea
86 c3p8cE_
not modelled
5.3
26
PDB header: protein bindingChain: E: PDB Molecule: probable protein brick1;PDBTitle: structure and control of the actin regulatory wave complex
87 d1t3ua_
not modelled
5.2
19
Fold: Cell division protein ZapA-likeSuperfamily: Cell division protein ZapA-likeFamily: Cell division protein ZapA-like
88 c3zeyZ_
not modelled
5.1
33
PDB header: ribosomeChain: Z: PDB Molecule: 40s ribosomal protein s33, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
89 c1t6fA_
not modelled
5.1
9
PDB header: cell cycleChain: A: PDB Molecule: geminin;PDBTitle: crystal structure of the coiled-coil dimerization motif of2 geminin
90 c4ypiG_
not modelled
5.1
23
PDB header: rna binding proteinChain: G: PDB Molecule: polymerase cofactor vp35;PDBTitle: structure of ebola virus nucleoprotein n-terminal fragment bound to a2 peptide derived from ebola vp35
91 c4ypiF_
not modelled
5.1
23
PDB header: rna binding proteinChain: F: PDB Molecule: polymerase cofactor vp35;PDBTitle: structure of ebola virus nucleoprotein n-terminal fragment bound to a2 peptide derived from ebola vp35