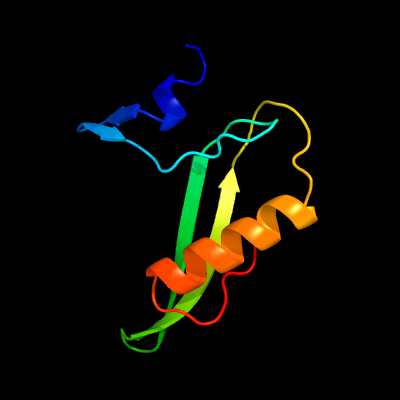
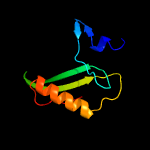
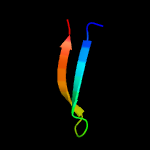
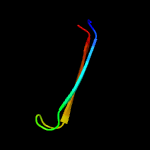
1 c5uujA_
98.0
15
PDB header: hydrolaseChain: A: PDB Molecule: alkz;PDBTitle: streptomyces sahachiroi dna glycosylase alkz
2 d2ae8a2
47.4
25
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase
3 d1rhya2
45.6
35
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase
4 d2f1da2
41.7
25
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase
5 c4icxA_
39.5
22
PDB header: transport proteinChain: A: PDB Molecule: centrosomal protein of 120 kda;PDBTitle: n-terminal c2 domain of human cep120
6 c2f1dP_
37.1
25
PDB header: lyaseChain: P: PDB Molecule: imidazoleglycerol-phosphate dehydratase 1;PDBTitle: x-ray structure of imidazoleglycerol-phosphate dehydratase
7 c1rhyB_
37.1
35
PDB header: lyaseChain: B: PDB Molecule: imidazole glycerol phosphate dehydratase;PDBTitle: crystal structure of imidazole glycerol phosphate dehydratase
8 c2ae8C_
36.1
25
PDB header: lyaseChain: C: PDB Molecule: imidazoleglycerol-phosphate dehydratase;PDBTitle: crystal structure of imidazoleglycerol-phosphate dehydratase from2 staphylococcus aureus subsp. aureus n315
9 c5dnlA_
31.8
30
PDB header: lyaseChain: A: PDB Molecule: imidazoleglycerol-phosphate dehydratase;PDBTitle: crystal structure of igpd from pyrococcus furiosus in complex with2 (s)-c348
10 c4lomA_
30.8
35
PDB header: lyaseChain: A: PDB Molecule: imidazoleglycerol-phosphate dehydratase;PDBTitle: crystal structure of mycobacterium tuberculosis hisb in complex with2 its substrate
11 c6ezmL_
29.1
35
PDB header: lyaseChain: L: PDB Molecule: imidazoleglycerol-phosphate dehydratase;PDBTitle: imidazoleglycerol-phosphate dehydratase from saccharomyces cerevisiae
12 c2aj6A_
27.5
14
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein mw0638;PDBTitle: crystal structure of a putative gnat family acetyltransferase (mw0638)2 from staphylococcus aureus subsp. aureus at 1.63 a resolution
13 c6fwhH_
24.0
35
PDB header: lyaseChain: H: PDB Molecule: imidazoleglycerol-phosphate dehydratase;PDBTitle: acanthamoeba igpd in complex with r-c348 to 1.7a resolution
14 d2hfqa1
22.5
17
Fold: NE1680-likeSuperfamily: NE1680-likeFamily: NE1680-like
15 c2hfqA_
22.5
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: nmr structure of protein ne1680 from nitrosomonas europaea:2 northeast structural genomics consortium target net5
16 d2aj6a1
22.2
14
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT
17 d1vkoa1
20.7
10
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
18 c5i0cA_
20.7
6
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein yjdj;PDBTitle: crystal structure of predicted acyltransferase yjdj with acyl-coa n-2 acyltransferase domain from escherichia coli str. k-12
19 d2ghvc1
19.1
19
Fold: SARS receptor-binding domain-likeSuperfamily: SARS receptor-binding domain-likeFamily: SARS receptor-binding domain-like
20 c4rrcA_
17.4
20
PDB header: ligaseChain: A: PDB Molecule: probable threonine--trna ligase 2;PDBTitle: n-terminal editing domain of threonyl-trna synthetase from aeropyrum2 pernix with l-thr3aa (snapshot 3)
21 c1j2oA_
not modelled
17.1
38
PDB header: metal binding proteinChain: A: PDB Molecule: fusion of rhombotin-2 and lim domain-bindingPDBTitle: structure of flin2, a complex containing the n-terminal lim2 domain of lmo2 and ldb1-lid
22 d1p1ja1
not modelled
16.9
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
23 c5gyqA_
not modelled
16.3
19
PDB header: viral proteinChain: A: PDB Molecule: spike glycoprotein;PDBTitle: putative receptor-binding domain of bat-derived coronavirus hku9 spike2 protein
24 c5xgrF_
not modelled
15.4
19
PDB header: viral proteinChain: F: PDB Molecule: spike protein s1;PDBTitle: structure of the s1 subunit c-terminal domain from bat-derived2 coronavirus hku5 spike protein
25 c4qzvB_
not modelled
13.9
11
PDB header: hydrolase/viral proteinChain: B: PDB Molecule: spike protein s1;PDBTitle: bat-derived coronavirus hku4 uses mers-cov receptor human cd26 for2 cell entry
26 c6hu9u_
not modelled
11.2
32
PDB header: oxidoreductase/electron transportChain: U: PDB Molecule: cytochrome b-c1 complex subunit 10;PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
27 c2r98A_
not modelled
10.5
17
PDB header: transferaseChain: A: PDB Molecule: putative acetylglutamate synthase;PDBTitle: crystal structure of n-acetylglutamate synthase (selenomet2 substituted) from neisseria gonorrhoeae
28 c3sciE_
not modelled
9.6
20
PDB header: hydrolase/viral proteinChain: E: PDB Molecule: spike glycoprotein;PDBTitle: crystal structure of spike protein receptor-binding domain from a2 predicted sars coronavirus human strain complexed with human receptor3 ace2
29 c3lodA_
not modelled
9.4
13
PDB header: transferaseChain: A: PDB Molecule: putative acyl-coa n-acyltransferase;PDBTitle: the crystal structure of the putative acyl-coa n-acyltransferase from2 klebsiella pneumoniae subsp.pneumoniae mgh 78578
30 c1p1hD_
not modelled
8.8
11
PDB header: isomeraseChain: D: PDB Molecule: inositol-3-phosphate synthase;PDBTitle: crystal structure of the 1l-myo-inositol/nad+ complex
31 c4kqzA_
not modelled
8.8
11
PDB header: viral proteinChain: A: PDB Molecule: s protein;PDBTitle: structure of the receptor binding domain (rbd) of mers-cov spike
32 c2atmA_
not modelled
7.2
27
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronoglucosaminidase;PDBTitle: crystal structure of the recombinant allergen ves v 2
33 c1w18A_
not modelled
7.1
25
PDB header: transferaseChain: A: PDB Molecule: levansucrase;PDBTitle: crystal structure of levansucrase from gluconacetobacter2 diazotrophicus
34 d1bwva2
not modelled
6.9
28
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase
35 c2hl2A_
not modelled
6.8
22
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase;PDBTitle: crystal structure of the editing domain of threonyl-trna2 synthetase from pyrococcus abyssi in complex with an3 analog of seryladenylate
36 c3d8pB_
not modelled
6.7
7
PDB header: transferaseChain: B: PDB Molecule: acetyltransferase of gnat family;PDBTitle: crystal structure of acetyltransferase of gnat family (np_373092.1)2 from staphylococcus aureus mu50 at 2.20 a resolution
37 d1fcqa_
not modelled
6.2
27
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Bee venom hyaluronidase
38 c2q0yA_
not modelled
6.1
9
PDB header: transferaseChain: A: PDB Molecule: gcn5-related n-acetyltransferase;PDBTitle: crystal structure of gcn5-related n-acetyltransferase (yp_295895.1)2 from ralstonia eutropha jmp134 at 1.80 a resolution
39 c2eemA_
not modelled
6.1
54
PDB header: antibioticChain: A: PDB Molecule: mytilin-b;PDBTitle: solution structure of the synthetic mytilin
40 c1fcuA_
not modelled
5.9
27
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronoglucosaminidase;PDBTitle: crystal structure (trigonal) of bee venom hyaluronidase
41 c2vzzA_
not modelled
5.9
13
PDB header: transferaseChain: A: PDB Molecule: rv0802c;PDBTitle: crystal structure of rv0802c from mycobacterium2 tuberculosis in complex with succinyl-coa
42 c2pe4A_
not modelled
5.7
18
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronidase-1;PDBTitle: structure of human hyaluronidase 1, a hyaluronan hydrolyzing enzyme2 involved in tumor growth and angiogenesis
43 c1vkoA_
not modelled
5.7
9
PDB header: isomeraseChain: A: PDB Molecule: inositol-3-phosphate synthase;PDBTitle: crystal structure of inositol-3-phosphate synthase (ce21227) from2 caenorhabditis elegans at 2.30 a resolution
44 d1gk8a2
not modelled
5.7
24
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase
45 c5wvxA_
not modelled
5.5
25
PDB header: hydrolase inhibitorChain: A: PDB Molecule: trypsin/chymotrypsin inhibitor;PDBTitle: crystal structure of bifunctional kunitz type trypsin /amylase2 inhibitor (amtin) from the tubers of alocasia macrorrhiza
46 c4rasC_
not modelled
5.4
13
PDB header: oxidoreductaseChain: C: PDB Molecule: oxidoreductase, nad-binding/iron-sulfur cluster-bindingPDBTitle: reductive dehalogenase structure suggests a mechanism for b12-2 dependent dehalogenation
47 c5ys3B_
not modelled
5.4
8
PDB header: transport proteinChain: B: PDB Molecule: succinate-acetate permease;PDBTitle: 1.8 angstrom crystal structure of succinate-acetate permease from2 citrobacter koseri
48 c2wpwA_
not modelled
5.3
12
PDB header: transferaseChain: A: PDB Molecule: orf14;PDBTitle: tandem gnat protein from the clavulanic acid biosynthesis pathway2 (without accoa)
49 c2z5cA_
not modelled
5.1
37
PDB header: chaperone/hydrolaseChain: A: PDB Molecule: protein ypl144w;PDBTitle: crystal structure of a novel chaperone complex for yeast 20s2 proteasome assembly