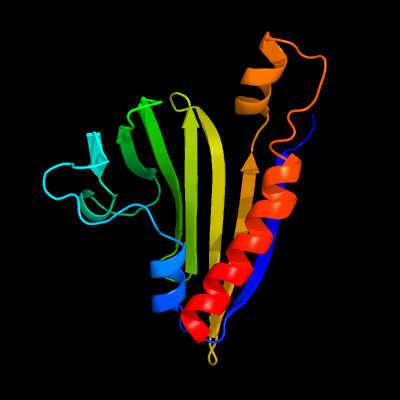
| 1 | c2m47A_
|
|
 |
99.9 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein cgl2373;
PDBTitle: solution nmr structure of the polyketide_cyc-like protein cgl2372 from2 corynebacterium glutamicum, northeast structural genomics consortium3 target cgr160
|
|
|
|
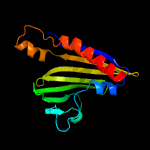
| 2 | c2kf2A_
|
|
 |
99.9 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative polyketide cyclase;
PDBTitle: solution nmr structure of of streptomyces coelicolor2 polyketide cyclase sco5315. northeast structural genomics3 consortium target rr365
|
|
|
|
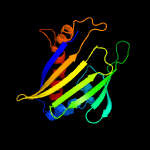
| 3 | d2d4ra1
|
|
 |
99.9 |
17 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:oligoketide cyclase/dehydrase-like |
|
|
|
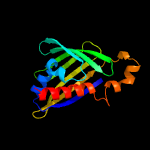
| 4 | d2rera1
|
|
 |
99.9 |
15 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:oligoketide cyclase/dehydrase-like |
|
|
|
| 5 | c3tfzB_
|
|
 |
99.9 |
17 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:cyclase;
PDBTitle: crystal structure of zhui aromatase/cyclase from streptomcyes sp.2 r1128
|
|
|
|
| 6 | c4xrwA_
|
|
 |
99.9 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:bexl;
PDBTitle: crystal structure of the di-domain aro/cyc bexl from the be-7585a2 biosynthetic pathway
|
|
|
|
| 7 | d2b79a1
|
|
 |
99.8 |
10 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Smu440-like |
|
|
|
| 8 | c5z8oA_
|
|
 |
99.8 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:cyclase/dehydrase;
PDBTitle: structural of start superfamily protein msmeg_0129 from mycobacterium2 smegmatis
|
|
|
|
| 9 | d1t17a_
|
|
 |
99.8 |
13 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:oligoketide cyclase/dehydrase-like |
|
|
|
| 10 | d2ns9a1
|
|
 |
99.8 |
10 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:CoxG-like |
|
|
|
| 11 | c5woxA_
|
|
 |
99.8 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: nmr solution structure of kany protein (ms6282) using two 4d-spectra
|
|
|
|
| 12 | c4xrtB_
|
|
 |
99.8 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:stfq aromatase/cyclase;
PDBTitle: crystal structure of the di-domain aro/cyc stfq from the steffimycin2 biosynthetic pathway
|
|
|
|
| 13 | d2pcsa1
|
|
 |
99.7 |
13 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:CoxG-like |
|
|
|
| 14 | c2lf2A_
|
|
 |
99.7 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the ahsa1-like protein chu_1110 from2 cytophaga hutchinsonii, northeast structural genomics consortium3 target chr152
|
|
|
|
| 15 | c2kczA_
|
|
 |
99.7 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein dr_a0006;
PDBTitle: solution nmr structure of the c-terminal domain of protein2 dr_a0006 from deinococcus radiodurans. northeast3 structural genomics consortium target drr147d
|
|
|
|
| 16 | c2lghA_
|
|
 |
99.7 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the ahsa1-like protein aha_2358 from2 aeromonas hydrophila refined with nh rdcs, northeast structural3 genomics consortium target ahr99.
|
|
|
|
| 17 | c3p9vA_
|
|
 |
99.7 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: high resolution crystal structure of protein maqu_3174 from2 marinobacter aquaeolei, northeast structural genomics consortium3 target mqr197
|
|
|
|
| 18 | d1xuva_
|
|
 |
99.7 |
16 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
|
| 19 | c2lcgA_
|
|
 |
99.7 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of protein rmet_5065 from ralstonia2 metallidurans, northeast structural genomics consortium target crr115
|
|
|
|
| 20 | c2l9pA_
|
|
 |
99.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of q5hli9 from staphylococcus epidermidis,2 northeast structural genomics consortium target ser147
|
|
|
|
| 21 | c2leqA_ |
|
not modelled |
99.7 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: chemical shift assignment and solution structure of chr145 from2 cytophaga hutchinsonii, northeast structural genomics consortium3 target chr145
|
|
|
| 22 | c2l8oA_ |
|
not modelled |
99.7 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of chr148 from cytophaga hutchinsonii, northeast2 structural genomics consortium target chr148
|
|
|
| 23 | c2le1A_ |
|
not modelled |
99.7 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of tfu_2981 from thermobifida fusca, northeast2 structural genomics consortium target tfr85a
|
|
|
| 24 | d1z94a1 |
|
not modelled |
99.7 |
14 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 25 | d1xfsa_ |
|
not modelled |
99.7 |
13 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 26 | c3uidA_ |
|
not modelled |
99.7 |
19 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of protein ms6760 from mycobacterium smegmatis
|
|
|
| 27 | c3pu2G_ |
|
not modelled |
99.7 |
16 |
PDB header:structure genomics, unknown function
Chain: G: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the q3j4m4_rhos4 protein from rhodobacter2 sphaeroides. northeast structural genomics consortium target rhr263.
|
|
|
| 28 | c2ldkA_ |
|
not modelled |
99.7 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of protein aaur_3427 from arthrobacter2 aurescens, northeast structural genomics consortium target aar96
|
|
|
| 29 | d2qpva1 |
|
not modelled |
99.7 |
14 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Atu1531-like |
|
|
| 30 | c2m89A_ |
|
not modelled |
99.7 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:aha1 domain protein;
PDBTitle: solution structure of the aha1 dimer from colwellia psychrerythraea
|
|
|
| 31 | c4n0gC_ |
|
not modelled |
99.7 |
15 |
PDB header:hydrolase/receptor
Chain: C: PDB Molecule:abscisic acid receptor pyl13;
PDBTitle: crystal structure of pyl13-pp2ca complex
|
|
|
| 32 | d3cnwa1 |
|
not modelled |
99.7 |
15 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Atu1531-like |
|
|
| 33 | c3q63F_ |
|
not modelled |
99.6 |
15 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:mll2253 protein;
PDBTitle: x-ray crystal structure of protein mll2253 from mesorhizobium loti,2 northeast structural genomics consortium target mlr404.
|
|
|
| 34 | c5vglA_ |
|
not modelled |
99.6 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:lachrymatory-factor synthase;
PDBTitle: crystal structure of lachrymatory factor synthase from allium cepa
|
|
|
| 35 | c4r7kA_ |
|
not modelled |
99.6 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein jhp0584;
PDBTitle: 1.88 angstrom resolution crystal structure of hypothetical protein2 jhp0584 from helicobacter pylori.
|
|
|
| 36 | d1xn5a_ |
|
not modelled |
99.6 |
17 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 37 | c3q6aH_ |
|
not modelled |
99.6 |
15 |
PDB header:structure genomics, unknown function
Chain: H: PDB Molecule:uncharacterized protein;
PDBTitle: x-ray crystal structure of the protein ssp2350 from staphylococcus2 saprophyticus, northeast structural genomics consortium target syr116
|
|
|
| 38 | c3otlA_ |
|
not modelled |
99.6 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: three-dimensional structure of the putative uncharacterized protein2 from rhizobium leguminosarum at the resolution 1.9a, northeast3 structural genomics consortium target rlr261
|
|
|
| 39 | c2nn5A_ |
|
not modelled |
99.6 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ef_2215;
PDBTitle: structure of conserved protein of unknown function ef2215 from2 enterococcus faecalis
|
|
|
| 40 | c3k90C_ |
|
not modelled |
99.6 |
17 |
PDB header:hormone receptor, hydrolase regulator
Chain: C: PDB Molecule:putative uncharacterized protein;
PDBTitle: the abscisic acid receptor pyr1 in complex with abscisic acid
|
|
|
| 41 | d2il5a1 |
|
not modelled |
99.6 |
10 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 42 | c3q64A_ |
|
not modelled |
99.6 |
15 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:mll3774 protein;
PDBTitle: x-ray crystal structure of protein mll3774 from mesorhizobium loti,2 northeast structural genomics consortium target mlr405.
|
|
|
| 43 | c3rd6A_ |
|
not modelled |
99.6 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:mll3558 protein;
PDBTitle: crystal structure of mll3558 protein from rhizobium loti. northeast2 structural genomics consortium target id mlr403
|
|
|
| 44 | c3p51A_ |
|
not modelled |
99.5 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: three-dimensional structure of protein q2y8n9_nitmu from nitrosospira2 multiformis, northeast structural genomics consortium target nmr118
|
|
|
| 45 | d2nn5a1 |
|
not modelled |
99.5 |
13 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 46 | c2lakA_ |
|
not modelled |
99.5 |
18 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:ahsa1-like protein rhe_ch02687;
PDBTitle: solution nmr structure of the ahsa1-like protein rhe_ch02687 (1-152)2 from rhizobium etli, northeast structural genomics consortium target3 rer242
|
|
|
| 47 | c3oquB_ |
|
not modelled |
99.5 |
10 |
PDB header:hormone receptor
Chain: B: PDB Molecule:abscisic acid receptor pyl9;
PDBTitle: crystal structure of native abscisic acid receptor pyl9 with aba
|
|
|
| 48 | d1xn6a_ |
|
not modelled |
99.5 |
12 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 49 | c4oicA_ |
|
not modelled |
99.5 |
17 |
PDB header:hormone receptor/hydrolase
Chain: A: PDB Molecule:bet v i allergen-like;
PDBTitle: crystal structrual of a soluble protein
|
|
|
| 50 | c3qtjA_ |
|
not modelled |
99.5 |
16 |
PDB header:hormone receptor
Chain: A: PDB Molecule:abscisic acid receptor pyl10;
PDBTitle: crystal strcuture of aba receptor pyl10 (apo)
|
|
|
| 51 | c3klxB_ |
|
not modelled |
99.5 |
15 |
PDB header:hormone receptor
Chain: B: PDB Molecule:f3n23.20 protein;
PDBTitle: crystal structure of native abscisic acid receptor pyl3
|
|
|
| 52 | c2kewA_ |
|
not modelled |
99.5 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yndb;
PDBTitle: the solution structure of bacillus subtilis sr211 start domain by nmr2 spectroscopy
|
|
|
| 53 | c2luzA_ |
|
not modelled |
99.5 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:calu16;
PDBTitle: solution nmr structure of calu16 from micromonospora echinospora,2 northeast structural genomics consortium (nesg) target mir12
|
|
|
| 54 | c3oh8A_ |
|
not modelled |
99.4 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate sugar epimerase (sula family);
PDBTitle: crystal structure of the nucleoside-diphosphate sugar epimerase from2 corynebacterium glutamicum. northeast structural genomics consortium3 target cgr91
|
|
|
| 55 | c5e4bB_ |
|
not modelled |
99.4 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:hydroxynitrile lyase;
PDBTitle: hydroxynitrile lyase from the fern davallia tyermanii in complex with2 (r)-mandelonitrile / benzaldehyde
|
|
|
| 56 | d2k5ga1 |
|
not modelled |
99.4 |
14 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 57 | c3kdiA_ |
|
not modelled |
99.4 |
18 |
PDB header:hormone receptor
Chain: A: PDB Molecule:putative uncharacterized protein at2g26040;
PDBTitle: structure of (+)-aba bound pyl2
|
|
|
| 58 | d3elia1 |
|
not modelled |
99.4 |
15 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 59 | c5ujvA_ |
|
not modelled |
99.4 |
16 |
PDB header:abscisic acid binding protein
Chain: A: PDB Molecule:pyr1;
PDBTitle: crystal structure of fepyr1 in complex with abscisic acid
|
|
|
| 60 | d1zxfa1 |
|
not modelled |
99.3 |
11 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 61 | d1x53a1 |
|
not modelled |
99.3 |
10 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:AHSA1 domain |
|
|
| 62 | c4igyB_ |
|
not modelled |
99.3 |
13 |
PDB header:allergen
Chain: B: PDB Molecule:kirola;
PDBTitle: crystal structure of kirola (act d 11) - triclinic form
|
|
|
| 63 | c5z4eA_ |
|
not modelled |
99.1 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:txno9;
PDBTitle: an anthrahydroquino-gama-pyrone synthase txn09
|
|
|
| 64 | c3ni8A_ |
|
not modelled |
99.1 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:pfc0360w protein;
PDBTitle: crystal structure of pfc0360w, an hsp90 activator from plasmodium2 falciparum
|
|
|
| 65 | d1ifva_ |
|
not modelled |
99.1 |
10 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
|
|
| 66 | c4rejA_ |
|
not modelled |
99.0 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:major latex-like protein;
PDBTitle: crystal structure of ginseng major latex-like protein 151 (glp) from2 panax ginseng. (crystal-3)
|
|
|
| 67 | c2k7hA_ |
|
not modelled |
99.0 |
13 |
PDB header:allergen
Chain: A: PDB Molecule:stress-induced protein sam22;
PDBTitle: nmr solution structure of soybean allergen gly m 4
|
|
|
| 68 | d1icxa_ |
|
not modelled |
99.0 |
10 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
|
|
| 69 | c2vq5B_ |
|
not modelled |
99.0 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:s-norcoclaurine synthase;
PDBTitle: x-ray structure of norcoclaurine synthase from thalictrum2 flavum in complex with dopamine and hydroxybenzaldehyde
|
|
|
| 70 | c3c0vC_ |
|
not modelled |
99.0 |
9 |
PDB header:plant protein
Chain: C: PDB Molecule:cytokinin-specific binding protein;
PDBTitle: crystal structure of cytokinin-specific binding protein in complex2 with cytokinin and ta6br12
|
|
|
| 71 | c3ie5A_ |
|
not modelled |
98.9 |
10 |
PDB header:plant protein, biosynthetic protein
Chain: A: PDB Molecule:phenolic oxidative coupling protein hyp-1;
PDBTitle: crystal structure of hyp-1 protein from hypericum perforatum (st2 john's wort) involved in hypericin biosynthesis
|
|
|
| 72 | d2bk0a1 |
|
not modelled |
98.9 |
16 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
|
|
| 73 | d1e09a_ |
|
not modelled |
98.9 |
11 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
|
|
| 74 | c3qszB_ |
|
not modelled |
98.9 |
16 |
PDB header:unknown function
Chain: B: PDB Molecule:star-related lipid transfer protein;
PDBTitle: crystal structure of the star-related lipid transfer protein (fragment2 25-204) from xanthomonas axonopodis at the resolution 2.4a, northeast3 structural genomics consortium target xar342
|
|
|
| 75 | d1xdfa1 |
|
not modelled |
98.9 |
13 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
|
|
| 76 | d1fm4a_ |
|
not modelled |
98.9 |
13 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
|
|
| 77 | c6gq9A_ |
|
not modelled |
98.8 |
10 |
PDB header:allergen
Chain: A: PDB Molecule:major allergen cor a 1.0401;
PDBTitle: solution structure of the hazel allergen cor a 1.0401
|
|
|
| 78 | d1qmra_ |
|
not modelled |
98.8 |
14 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
|
|
| 79 | c2i9yA_ |
|
not modelled |
98.8 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:major latex protein-like protein 28 or mlp-like
PDBTitle: solution structure of arabidopsis thaliana protein2 at1g70830, a member of the major latex protein family
|
|
|
| 80 | c3rwsA_ |
|
not modelled |
98.7 |
14 |
PDB header:plant protein
Chain: A: PDB Molecule:mtn13 protein;
PDBTitle: crystal structure of medicago truncatula nodulin 13 (mtn13) in complex2 with trans-zeatin
|
|
|
| 81 | d1txca1 |
|
not modelled |
98.6 |
12 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
|
|
| 82 | c2r55B_ |
|
not modelled |
98.0 |
7 |
PDB header:transport protein
Chain: B: PDB Molecule:star-related lipid transfer protein 5;
PDBTitle: human star-related lipid transfer protein 5
|
|
|
| 83 | c3fo5A_ |
|
not modelled |
98.0 |
12 |
PDB header:lipid transport
Chain: A: PDB Molecule:thioesterase, adipose associated, isoform bfit2;
PDBTitle: human start domain of acyl-coenzyme a thioesterase 11 (acot11)
|
|
|
| 84 | c3p0lC_ |
|
not modelled |
97.7 |
11 |
PDB header:transport protein
Chain: C: PDB Molecule:steroidogenic acute regulatory protein, mitochondrial;
PDBTitle: human steroidogenic acute regulatory protein
|
|
|
| 85 | d1em2a_ |
|
not modelled |
97.6 |
10 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:STAR domain |
|
|
| 86 | c2mouA_ |
|
not modelled |
97.2 |
5 |
PDB header:transport protein
Chain: A: PDB Molecule:star-related lipid transfer protein 6;
PDBTitle: solution structure of star-related lipid transfer domain protein 62 (stard6)
|
|
|
| 87 | c1jssB_ |
|
not modelled |
97.1 |
8 |
PDB header:lipid binding protein
Chain: B: PDB Molecule:cholesterol-regulated start protein 4;
PDBTitle: crystal structure of the mus musculus cholesterol-regulated2 start protein 4 (stard4).
|
|
|
| 88 | d1jssa_ |
|
not modelled |
96.9 |
9 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:STAR domain |
|
|
| 89 | d1vjha_ |
|
not modelled |
93.2 |
9 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Pathogenesis-related protein 10 (PR10)-like |
|
|
| 90 | d1ln1a_ |
|
not modelled |
92.3 |
10 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:STAR domain |
|
|
| 91 | c2e3rB_ |
|
not modelled |
87.7 |
11 |
PDB header:lipid transport
Chain: B: PDB Molecule:lipid-transfer protein cert;
PDBTitle: crystal structure of cert start domain in complex with c18-2 ceramide (p1)
|
|
|
| 92 | d2psoa1 |
|
not modelled |
86.9 |
13 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:STAR domain |
|
|
| 93 | c2lioA_ |
|
not modelled |
85.6 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of bfr322 from bacteroides fragilis, northeast2 structural genomics consortium target bfr322
|
|
|
| 94 | c2ejxA_ |
|
not modelled |
79.9 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:stk_08120;
PDBTitle: crystal structure of the hypothetical protein stk_08120 from2 sulfolobus tokodaii
|
|
|
| 95 | c3qrzC_ |
|
not modelled |
48.0 |
27 |
PDB header:hormone receptor
Chain: C: PDB Molecule:abscisic acid receptor pyl5;
PDBTitle: crystal structure of native abscisic acid receptor pyl5 at 2.62 angstrom
|
|
|
| 96 | c2lafA_ |
|
not modelled |
46.8 |
6 |
PDB header:membrane protein
Chain: A: PDB Molecule:lipoprotein 34;
PDBTitle: nmr solution structure of the n-terminal domain of the e. coli2 lipoprotein bamc
|
|
|
| 97 | c5ys0A_ |
|
not modelled |
22.6 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:membrane-anchored lipid-binding protein ysp2;
PDBTitle: crystal structure of the second starkin domain of lam2 in complex with2 ergosterol
|
|
|
| 98 | c2oqgA_ |
|
not modelled |
20.3 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:possible transcriptional regulator, arsr family protein;
PDBTitle: arsr-like transcriptional regulator from rhodococcus sp. rha1
|
|
|
| 99 | c6gqfC_ |
|
not modelled |
20.3 |
16 |
PDB header:lipid transport
Chain: C: PDB Molecule:gram domain-containing protein 1a;
PDBTitle: the structure of mouse astera (gramd1a) with 25-hydroxy cholesterol
|
|
|