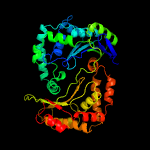
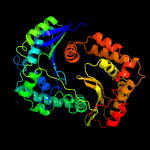
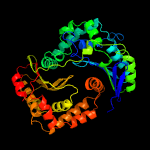
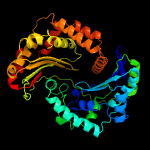
1 c1hr9D_
100.0
28
PDB header: hydrolaseChain: D: PDB Molecule: mitochondrial processing peptidase beta subunit;PDBTitle: yeast mitochondrial processing peptidase beta-e73q mutant2 complexed with malate dehydrogenase signal peptide
2 c1sqpA_
100.0
29
PDB header: oxidoreductaseChain: A: PDB Molecule: ubiquinol-cytochrome-c reductase complex core protein i,PDBTitle: crystal structure analysis of bovine bc1 with myxothiazol
3 c1nu1A_
100.0
29
PDB header: oxidoreductaseChain: A: PDB Molecule: ubiquinol-cytochrome c reductase complex core protein i,PDBTitle: crystal structure of mitochondrial cytochrome bc1 complexed with 2-2 nonyl-4-hydroxyquinoline n-oxide (nqno)
4 c1hr6C_
100.0
24
PDB header: hydrolaseChain: C: PDB Molecule: mitochondrial processing peptidase alpha subunit;PDBTitle: yeast mitochondrial processing peptidase
5 c3hdiA_
100.0
38
PDB header: hydrolaseChain: A: PDB Molecule: processing protease;PDBTitle: crystal structure of bacillus halodurans metallo peptidase
6 c5eufA_
100.0
25
PDB header: hydrolaseChain: A: PDB Molecule: protease;PDBTitle: the crystal structure of a protease from helicobacter pylori
7 c3eoqB_
100.0
30
PDB header: hydrolaseChain: B: PDB Molecule: putative zinc protease;PDBTitle: the crystal structure of putative zinc protease beta-2 subunit from thermus thermophilus hb8
8 c1l0lB_
100.0
21
PDB header: oxidoreductaseChain: B: PDB Molecule: ubiquinol-cytochrome c reductase complex core protein 2;PDBTitle: structure of bovine mitochondrial cytochrome bc1 complex with a bound2 fungicide famoxadone
9 c5hxkC_
100.0
29
PDB header: hydrolaseChain: C: PDB Molecule: zinc-dependent peptidase;PDBTitle: structure of ttha1265
10 c3amiB_
100.0
25
PDB header: hydrolaseChain: B: PDB Molecule: zinc peptidase;PDBTitle: the crystal structure of the m16b metallopeptidase subunit from2 sphingomonas sp. a1
11 c3gwbA_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: peptidase m16 inactive domain family protein;PDBTitle: crystal structure of peptidase m16 inactive domain from pseudomonas2 fluorescens. northeast structural genomics target plr293l
12 c3amjB_
100.0
24
PDB header: hydrolaseChain: B: PDB Molecule: zinc peptidase inactive subunit;PDBTitle: the crystal structure of the heterodimer of m16b peptidase from2 sphingomonas sp. a1
13 c4nnzB_
100.0
23
PDB header: hydrolaseChain: B: PDB Molecule: probable zinc protease;PDBTitle: subunit pa0372 of heterodimeric zinc protease pa0371-pa0372
14 c3cx5L_
100.0
19
PDB header: oxidoreductaseChain: L: PDB Molecule: cytochrome b-c1 complex subunit 1, mitochondrial;PDBTitle: structure of complex iii with bound cytochrome c in reduced state and2 definition of a minimal core interface for electron transfer.
15 c4xeaA_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: peptidase m16 domain protein;PDBTitle: crystal structure of putative m16-like peptidase from alicyclobacillus2 acidocaldarius
16 c6b05A_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: putative zinc protease;PDBTitle: the crystal structure of the ferredoxin protease fusc e83a mutant in2 complex with arabidopsis ferredoxin
17 c1q2lA_
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: protease iii;PDBTitle: crystal structure of pitrilysin
18 c3go9A_
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: insulinase family protease;PDBTitle: predicted insulinase family protease from yersinia pestis
19 c2wk3A_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: insulin degrading enzyme;PDBTitle: crystal structure of human insulin-degrading enzyme in2 complex with amyloid-beta (1-42)
20 c2jbuB_
100.0
17
PDB header: hydrolaseChain: B: PDB Molecule: insulin-degrading enzyme;PDBTitle: crystal structure of human insulin degrading enzyme complexed with co-2 purified peptides.
21 c3d3yA_
not modelled
100.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a conserved protein from enterococcus faecalis2 v583
22 c5cioA_
not modelled
100.0
21
PDB header: metal binding proteinChain: A: PDB Molecule: pyrroloquinoline quinone biosynthesis protein pqqf;PDBTitle: crystal structure of pqqf
23 c2fgeA_
not modelled
100.0
17
PDB header: hydrolase, plant proteinChain: A: PDB Molecule: zinc metalloprotease (insulinase family);PDBTitle: crystal structure of presequence protease prep from2 arabidopsis thaliana
24 c3s5mA_
not modelled
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: falcilysin;PDBTitle: crystal structures of falcilysin, a m16 metalloprotease from the2 malaria parasite plasmodium falciparum
25 c3s5kA_
not modelled
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: falcilysin;PDBTitle: crystal structures of falcilysin, a m16 metalloprotease from the2 malaria parasite plasmodium falciparum
26 c4l3tB_
not modelled
100.0
17
PDB header: hydrolaseChain: B: PDB Molecule: presequence protease, mitochondrial;PDBTitle: crystal structure of substrate-free human presequence protease
27 c3cxhM_
not modelled
100.0
18
PDB header: oxidoreductaseChain: M: PDB Molecule: cytochrome b-c1 complex subunit 2, mitochondrial;PDBTitle: structure of yeast complex iii with isoform-2 cytochrome c bound and2 definition of a minimal core interface for electron transfer.
28 d1ppja1
not modelled
100.0
34
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
29 d1hr6b1
not modelled
100.0
35
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
30 d1bcca1
not modelled
100.0
33
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
31 d1hr6a1
not modelled
100.0
24
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
32 d1q2la4
not modelled
100.0
24
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
33 d1ppjb1
not modelled
100.0
21
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
34 d1bccb1
not modelled
100.0
21
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
35 d3cx5a1
not modelled
100.0
21
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
36 d2fgea4
not modelled
100.0
21
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
37 d3cx5b1
not modelled
100.0
22
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
38 d1hr6a2
not modelled
100.0
26
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
39 d1ppjb2
not modelled
100.0
23
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
40 d1hr6b2
not modelled
99.9
20
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
41 d1bcca2
not modelled
99.9
22
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
42 c3ivlA_
not modelled
99.9
17
PDB header: hydrolaseChain: A: PDB Molecule: putative zinc protease;PDBTitle: the crystal structure of the inactive peptidase domain of a putative2 zinc protease from bordetella parapertussis to 2.2a
43 d1bccb2
not modelled
99.9
21
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
44 d1ppja2
not modelled
99.9
23
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
45 d1q2la1
not modelled
99.9
12
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
46 d3cx5a2
not modelled
99.9
18
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
47 d2fgea2
not modelled
99.9
12
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
48 d2fgea3
not modelled
99.6
12
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
49 d2fgea1
not modelled
99.5
10
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
50 d1q2la2
not modelled
99.5
10
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
51 d1q2la3
not modelled
98.8
10
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
52 d1nsha_
not modelled
31.9
14
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins
53 c6cy1B_
not modelled
28.5
10
PDB header: signaling proteinChain: B: PDB Molecule: signal recognition particle receptor ftsy;PDBTitle: crystal structure of signal recognition particle receptor ftsy from2 elizabethkingia anophelis
54 c2rgiA_
not modelled
24.2
11
PDB header: metal binding proteinChain: A: PDB Molecule: protein s100-a2;PDBTitle: crystal structure of ca2+-free s100a2 at 1.6 a resolution
55 d1qlsa_
not modelled
23.5
11
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins
56 c1qapA_
not modelled
20.7
16
PDB header: glycosyltransferaseChain: A: PDB Molecule: quinolinic acid phosphoribosyltransferase;PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid
57 c5yhvA_
not modelled
20.5
13
PDB header: transferaseChain: A: PDB Molecule: aminotransferase;PDBTitle: crystal structure of an aminotransferase from mycobacterium2 tuberculosis
58 d1zavz1
not modelled
20.4
29
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
59 c1zawZ_
not modelled
20.4
29
PDB header: structural proteinChain: Z: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
60 c1zavZ_
not modelled
20.4
29
PDB header: structural proteinChain: Z: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
61 d2azeb1
not modelled
20.3
21
Fold: E2F-DP heterodimerization regionSuperfamily: E2F-DP heterodimerization regionFamily: E2F dimerization segment
62 c1zawX_
not modelled
19.8
29
PDB header: structural proteinChain: X: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
63 c1zawY_
not modelled
19.8
29
PDB header: structural proteinChain: Y: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
64 c2y5iF_
not modelled
18.5
12
PDB header: metal-binding proteinChain: F: PDB Molecule: s100 calcium binding protein z;PDBTitle: s100z from zebrafish in complex with calcium
65 c5tuvB_
not modelled
18.5
27
PDB header: transcriptionChain: B: PDB Molecule: transcription factor e2f5;PDBTitle: crystal structure of the e2f5-dp1-p107 ternary complex
66 c3tcmB_
not modelled
18.2
9
PDB header: transferaseChain: B: PDB Molecule: alanine aminotransferase 2;PDBTitle: crystal structure of alanine aminotransferase from hordeum vulgare
67 c4ix9A_
not modelled
18.2
7
PDB header: hydrolaseChain: A: PDB Molecule: v-type proton atpase subunit f;PDBTitle: crystal structure of subunit f of v-atpase from s. cerevisiae
68 c1zavU_
not modelled
17.9
28
PDB header: structural proteinChain: U: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
69 c1zaxU_
not modelled
17.9
28
PDB header: structural proteinChain: U: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
70 d1zavu1
not modelled
17.9
28
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
71 c1zaxV_
not modelled
17.9
28
PDB header: structural proteinChain: V: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
72 c1zaxW_
not modelled
17.9
28
PDB header: structural proteinChain: W: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
73 c1zavV_
not modelled
17.9
28
PDB header: structural proteinChain: V: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
74 c1zavW_
not modelled
17.9
28
PDB header: structural proteinChain: W: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
75 c1zavX_
not modelled
17.7
28
PDB header: structural proteinChain: X: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
76 c1zavY_
not modelled
17.7
28
PDB header: structural proteinChain: Y: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
77 c1zaxY_
not modelled
17.7
28
PDB header: structural proteinChain: Y: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
78 c1zaxX_
not modelled
17.7
28
PDB header: structural proteinChain: X: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
79 d1b5pa_
not modelled
17.5
22
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
80 c1dd3D_
not modelled
17.3
28
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
81 c1dd3C_
not modelled
17.3
28
PDB header: ribosomeChain: C: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
82 c3pajA_
not modelled
16.3
13
PDB header: transferaseChain: A: PDB Molecule: nicotinate-nucleotide pyrophosphorylase, carboxylating;PDBTitle: 2.00 angstrom resolution crystal structure of a quinolinate2 phosphoribosyltransferase from vibrio cholerae o1 biovar eltor str.3 n16961
83 d1e8aa_
not modelled
15.5
5
Fold: EF Hand-likeSuperfamily: EF-handFamily: S100 proteins
84 c3ihjA_
not modelled
15.2
11
PDB header: transferaseChain: A: PDB Molecule: alanine aminotransferase 2;PDBTitle: human alanine aminotransferase 2 in complex with plp
85 d1c7na_
not modelled
15.1
6
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like
86 c4drwC_
not modelled
14.9
10
PDB header: exocytosis/protein bindingChain: C: PDB Molecule: protein s100-a10/annexin a2 chimeric protein;PDBTitle: crystal structure of the ternary complex between s100a10, an annexin2 a2 n-terminal peptide and an ahnak peptide
87 d1cr6a2
not modelled
14.6
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase
88 c1zawU_
not modelled
14.6
29
PDB header: structural proteinChain: U: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
89 c1zawW_
not modelled
14.6
29
PDB header: structural proteinChain: W: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
90 c1zawV_
not modelled
14.6
29
PDB header: structural proteinChain: V: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
91 c3gnnA_
not modelled
14.6
18
PDB header: transferaseChain: A: PDB Molecule: nicotinate-nucleotide pyrophosphorylase;PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 burkholderi pseudomallei
92 c5wmiA_
not modelled
14.6
13
PDB header: transferaseChain: A: PDB Molecule: bifunctional aspartate aminotransferase andPDBTitle: arabidopsis thaliana prephenate aminotransferase mutant- t84v
93 c1yhpA_
not modelled
14.5
15
PDB header: cell adhesionChain: A: PDB Molecule: calcium-dependent cell adhesion molecule-1;PDBTitle: solution structure of ca2+-free ddcad-1
94 c1x1oC_
not modelled
14.4
18
PDB header: transferaseChain: C: PDB Molecule: nicotinate-nucleotide pyrophosphorylase;PDBTitle: crystal structure of project id tt0268 from thermus thermophilus hb8
95 c1zaxZ_
not modelled
14.3
29
PDB header: structural proteinChain: Z: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
96 c3ff5B_
not modelled
14.0
21
PDB header: protein transportChain: B: PDB Molecule: peroxisomal biogenesis factor 14;PDBTitle: crystal structure of the conserved n-terminal domain of the2 peroxisomal matrix-protein-import receptor, pex14p
97 d1dd4c_
not modelled
13.7
28
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
98 c4f4fB_
not modelled
13.3
18
PDB header: lyaseChain: B: PDB Molecule: threonine synthase;PDBTitle: x-ray crystal structure of plp bound threonine synthase from brucella2 melitensis
99 d1d2fa_
not modelled
13.3
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like