1 c5o5jO_
100.0
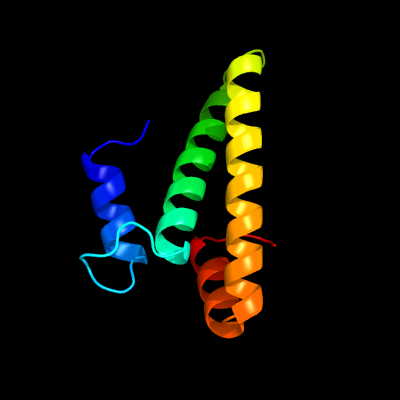
89
PDB header: ribosomeChain: O: PDB Molecule: 30s ribosomal protein s15;PDBTitle: structure of the 30s small ribosomal subunit from mycobacterium2 smegmatis
2 d1vs5o1
100.0
52
Fold: S15/NS1 RNA-binding domainSuperfamily: S15/NS1 RNA-binding domainFamily: Ribosomal protein S15
3 d1g1xb_
100.0
56
Fold: S15/NS1 RNA-binding domainSuperfamily: S15/NS1 RNA-binding domainFamily: Ribosomal protein S15
4 d1a32a_
100.0
61
Fold: S15/NS1 RNA-binding domainSuperfamily: S15/NS1 RNA-binding domainFamily: Ribosomal protein S15
5 c3ulwA_
100.0
51
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s15;PDBTitle: 30s ribosomal protein s15 from campylobacter jejuni
6 d1kuqa_
100.0
57
Fold: S15/NS1 RNA-binding domainSuperfamily: S15/NS1 RNA-binding domainFamily: Ribosomal protein S15
7 c3j6vO_
100.0
26
PDB header: ribosomeChain: O: PDB Molecule: 28s ribosomal protein s15, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
8 c3bbnO_
99.9
39
PDB header: ribosomeChain: O: PDB Molecule: ribosomal protein s15;PDBTitle: homology model for the spinach chloroplast 30s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome.
9 c2xzmO_
99.2
19
PDB header: ribosomeChain: O: PDB Molecule: rps13e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
10 c5xyiN_
99.1
27
PDB header: ribosomeChain: N: PDB Molecule: 40s ribosomal protein s13, putative;PDBTitle: small subunit of trichomonas vaginalis ribosome
11 c3izbO_
99.1
14
PDB header: ribosomeChain: O: PDB Molecule: 40s ribosomal protein rps13 (s15p);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
12 c3u5cN_
98.8
26
PDB header: ribosomeChain: N: PDB Molecule: 40s ribosomal protein s13;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome a
13 c1yshE_
97.9
25
PDB header: structural protein/rnaChain: E: PDB Molecule: 40s ribosomal protein s13;PDBTitle: localization and dynamic behavior of ribosomal protein l30e
14 c3zeyG_
97.8
28
PDB header: ribosomeChain: G: PDB Molecule: 40s ribosomal protein s13, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
15 c3j20Q_
96.2
43
PDB header: ribosomeChain: Q: PDB Molecule: 30s ribosomal protein s15p/s13e;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
16 c1s1hO_
96.0
19
PDB header: ribosomeChain: O: PDB Molecule: 40s ribosomal protein s13;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1h, contains 40s subunit. the 60s4 ribosomal subunit is in file 1s1i.
17 c4ar9A_
43.8
30
PDB header: hydrolaseChain: A: PDB Molecule: collagenase colt;PDBTitle: crystal structure of the peptidase domain of collagenase t2 from clostridium tetani at 1.69 angstrom resolution.
18 c4ar1A_
38.3
26
PDB header: hydrolaseChain: A: PDB Molecule: colh protein;PDBTitle: crystal structure of the peptidase domain of collagenase h from2 clostridium histolyticum at 2.01 angstrom resolution.
19 d1y0ua_
32.3
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators
20 c2y50A_
26.1
30
PDB header: hydrolaseChain: A: PDB Molecule: collagenase;PDBTitle: crystal structure of collagenase g from clostridium2 histolyticum at 2.80 angstrom resolution
21 c5ajaC_
not modelled
22.4
38
PDB header: viral proteinChain: C: PDB Molecule: sam domain and hd domain-containing protein;PDBTitle: crystal structure of mandrill samhd1 (amino acid residues 1-114)2 bound to vpx isolated from mandrill and human dcaf1 (amino3 acid residues 1058-1396)
22 c5dukA_
not modelled
21.7
28
PDB header: transcriptionChain: A: PDB Molecule: putative dna binding protein;PDBTitle: n-terminal structure of putative dna binding transcription factor from2 thermoplasmatales archaeon scgc ab-539-n05
23 c2ke4A_
not modelled
21.5
26
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
24 d1sxjd1
not modelled
21.0
15
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: DNA polymerase III clamp loader subunits, C-terminal domain
25 c2zvnF_
not modelled
17.6
11
PDB header: signaling protein/transcriptionChain: F: PDB Molecule: nf-kappa-b essential modulator;PDBTitle: nemo cozi domain incomplex with diubiquitin in p212121 space group
26 c3uosH_
not modelled
15.2
25
PDB header: ribosomeChain: H: PDB Molecule: 50s ribosomal protein l10;PDBTitle: crystal structure of release factor rf3 trapped in the gtp state on a2 rotated conformation of the ribosome (without viomycin)
27 c5frgA_
not modelled
13.7
21
PDB header: protein bindingChain: A: PDB Molecule: formin-binding protein 1-like;PDBTitle: the nmr structure of the cdc42-interacting region of toca1
28 c2lhuA_
not modelled
13.5
14
PDB header: structural proteinChain: A: PDB Molecule: mybpc3 protein;PDBTitle: structural insight into the unique cardiac myosin binding protein-c2 motif: a partially folded domain
29 c3f1iH_
not modelled
11.3
17
PDB header: protein bindingChain: H: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: human escrt-0 core complex
30 c4kmfA_
not modelled
11.2
19
PDB header: transferase/dnaChain: A: PDB Molecule: interferon-inducible and double-stranded-dependent eif-PDBTitle: crystal structure of zalpha domain from carassius auratus pkz in2 complex with z-dna
31 c2vogB_
not modelled
10.8
67
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-modifying factor;PDBTitle: structure of mouse a1 bound to the bmf bh3-domain
32 c4wyhA_
not modelled
10.7
35
PDB header: replicationChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of prix from the hyperthermophilic archaeon2 sulfolobus solfataricus
33 c2hjqA_
not modelled
10.4
21
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein yqbf;PDBTitle: nmr structure of bacillus subtilis protein yqbf, northeast2 structural genomics target sr449
34 c2v4hA_
not modelled
9.8
11
PDB header: transcriptionChain: A: PDB Molecule: nf-kappa-b essential modulator;PDBTitle: nemo cc2-lz domain - 1d5 darpin complex
35 c4lb6B_
not modelled
9.6
16
PDB header: transferase/dnaChain: B: PDB Molecule: protein kinase containing z-dna binding domains;PDBTitle: crystal structure of pkz zalpha in complex with ds(cg)6 (tetragonal2 form)
36 c3nqwB_
not modelled
8.9
13
PDB header: hydrolaseChain: B: PDB Molecule: cg11900;PDBTitle: a metazoan ortholog of spot hydrolyzes ppgpp and plays a role in2 starvation responses
37 c1ye9E_
not modelled
8.7
18
PDB header: oxidoreductaseChain: E: PDB Molecule: catalase hpii;PDBTitle: crystal structure of proteolytically truncated catalase2 hpii from e. coli
38 c5tzqD_
not modelled
8.5
67
PDB header: apoptosisChain: D: PDB Molecule: bcl-2-modifying factor;PDBTitle: crystal structure of fpv039:bmf bh3 complex
39 c5tzqC_
not modelled
8.5
67
PDB header: apoptosisChain: C: PDB Molecule: bcl-2-modifying factor;PDBTitle: crystal structure of fpv039:bmf bh3 complex
40 c2ymyB_
not modelled
8.3
83
PDB header: apoptosisChain: B: PDB Molecule: ras association domain-containing protein 5;PDBTitle: structure of the murine nore1-sarah domain
41 c2khnA_
not modelled
8.3
17
PDB header: signaling proteinChain: A: PDB Molecule: intersectin-1;PDBTitle: nmr solution structure of the eh 1 domain from human2 intersectin-1 protein. northeast structural genomics3 consortium target hr3646e.
42 c3rj1Q_
not modelled
8.2
13
PDB header: transcriptionChain: Q: PDB Molecule: mediator of rna polymerase ii transcription subunit 8;PDBTitle: architecture of the mediator head module
43 d2jdia1
not modelled
8.1
13
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase
44 d3bj1b1
not modelled
7.3
12
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins
45 c2f5xC_
not modelled
7.3
17
PDB header: transport proteinChain: C: PDB Molecule: bugd;PDBTitle: structure of periplasmic binding protein bugd
46 c3cuoB_
not modelled
6.9
20
PDB header: transcription regulatorChain: B: PDB Molecule: uncharacterized hth-type transcriptional regulator ygav;PDBTitle: crystal structure of the predicted dna-binding transcriptional2 regulator from e. coli
47 c4bj5A_
not modelled
6.7
35
PDB header: transcriptionChain: A: PDB Molecule: protein rif2;PDBTitle: crystal structure of rif2 in complex with the c-terminal domain of2 rap1 (rap1-rct)
48 c5d1vB_
not modelled
6.7
9
PDB header: oxygen transportChain: B: PDB Molecule: globin;PDBTitle: crystal structure and thermal stability of hemoglobin from2 thermophilic phototrophic bacterium chloroflexus aurantiacus
49 d1sxjb1
not modelled
6.6
19
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: DNA polymerase III clamp loader subunits, C-terminal domain
50 c3qkuC_
not modelled
6.6
16
PDB header: replicationChain: C: PDB Molecule: dna double-strand break repair protein mre11;PDBTitle: mre11 rad50 binding domain in complex with rad50 and amp-pnp
51 d1fx0a1
not modelled
6.4
10
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase
52 d3crda_
not modelled
6.4
8
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD
53 d2f43a1
not modelled
6.2
13
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase
54 d1biaa1
not modelled
6.2
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like
55 c3katA_
not modelled
5.9
20
PDB header: apoptosisChain: A: PDB Molecule: nacht, lrr and pyd domains-containing protein 1;PDBTitle: crystal structure of the card domain of the human nlrp1 protein,2 northeast structural genomics consortium target hr3486e
56 d2zjrw1
not modelled
5.6
27
Fold: Ribosomal protein L30p/L7eSuperfamily: Ribosomal protein L30p/L7eFamily: Ribosomal protein L30p/L7e
57 d1spgb_
not modelled
5.5
24
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins
58 c1m7lA_
not modelled
5.5
21
PDB header: sugar binding proteinChain: A: PDB Molecule: pulmonary surfactant-associated protein d;PDBTitle: solution structure of the coiled-coil trimerization domain2 from lung surfactant protein d
59 c3qkrC_
not modelled
5.5
16
PDB header: replicationChain: C: PDB Molecule: dna double-strand break repair protein mre11;PDBTitle: mre11 rad50 binding domain bound to rad50
60 c3ac6A_
not modelled
5.3
14
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylformylglycinamidine synthase 2;PDBTitle: crystal structure of purl from thermus thermophilus
61 c2b19A_
not modelled
5.3
50
PDB header: neuropeptideChain: A: PDB Molecule: neuropeptide k;PDBTitle: solution structure of mammalian tachykinin peptide,2 neuropeptide k
62 d1gyza_
not modelled
5.2
10
Fold: PABP domain-likeSuperfamily: Ribosomal protein L20Family: Ribosomal protein L20
63 c2nz7A_
not modelled
5.2
12
PDB header: apoptosisChain: A: PDB Molecule: caspase recruitment domain-containing protein 4;PDBTitle: crystal structure analysis of caspase-recruitment domain2 (card) of nod1
64 d2jxca1
not modelled
5.2
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)
65 c3j3vY_
not modelled
5.2
33
PDB header: ribosomeChain: Y: PDB Molecule: 50s ribosomal protein l30;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
66 c3m20A_
not modelled
5.1
23
PDB header: isomeraseChain: A: PDB Molecule: 4-oxalocrotonate tautomerase, putative;PDBTitle: crystal structure of dmpi from archaeoglobus fulgidus determined to2 2.37 angstroms resolution
67 d2b7ea1
not modelled
5.1
36
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain
68 c3piwA_
not modelled
5.1
14
PDB header: cytokineChain: A: PDB Molecule: type i interferon 2;PDBTitle: zebrafish interferon 2
69 c5o60a_
not modelled
5.1
27
PDB header: ribosomeChain: A: PDB Molecule: 23s rrna;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
70 c6dlcA_
not modelled
5.1
13
PDB header: de novo proteinChain: A: PDB Molecule: designed protein dhd1:234_a;PDBTitle: designed protein dhd1:234_a, designed protein dhd1:234_b
71 d1skyb1
not modelled
5.1
9
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase
72 c2l4aA_
not modelled
5.1
21
PDB header: dna binding proteinChain: A: PDB Molecule: leucine responsive regulatory protein;PDBTitle: nmr structure of the dna-binding domain of e.coli lrp
73 d1x4ta1
not modelled
5.0
20
Fold: Long alpha-hairpinSuperfamily: ISY1 domain-likeFamily: ISY1 N-terminal domain-like