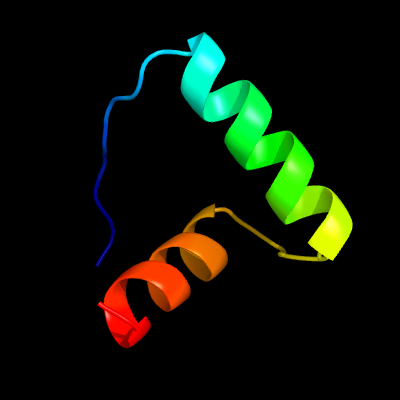
| 1 | d2bj7a1
|
|
 |
97.7 |
21 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
|
|
|
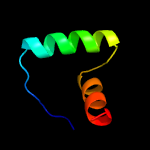
| 2 | c2bj3D_
|
|
 |
97.6 |
21 |
PDB header:transcription
Chain: D: PDB Molecule:nickel responsive regulator;
PDBTitle: nikr-apo
|
|
|
|
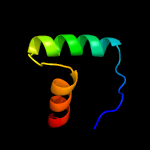
| 3 | d2hzaa1
|
|
 |
97.5 |
29 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
|
|
|
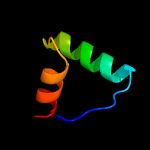
| 4 | c1q5vB_
|
|
 |
97.5 |
26 |
PDB header:transcription
Chain: B: PDB Molecule:nickel responsive regulator;
PDBTitle: apo-nikr
|
|
|
|
| 5 | d2hzab1
|
|
 |
97.4 |
29 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
|
|
|
| 6 | c2ca9B_
|
|
 |
97.4 |
32 |
PDB header:transcription
Chain: B: PDB Molecule:putative nickel-responsive regulator;
PDBTitle: apo-nikr from helicobacter pylori in closed trans-2 conformation
|
|
|
|
| 7 | c5cegC_
|
|
 |
94.5 |
19 |
PDB header:toxin
Chain: C: PDB Molecule:addiction module antidote protein, copg/arc/metj family;
PDBTitle: x-ray structure of toxin/anti-toxin complex from mesorhizobium2 opportunistum
|
|
|
|
| 8 | c5yrzC_
|
|
 |
93.8 |
26 |
PDB header:antitoxin/hydrolase
Chain: C: PDB Molecule:hicb;
PDBTitle: toxin-antitoxin complex from streptococcus pneumoniae
|
|
|
|
| 9 | c3kxeD_
|
|
 |
93.3 |
22 |
PDB header:protein binding
Chain: D: PDB Molecule:antitoxin protein pard-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
|
|
|
|
| 10 | c6g1nB_
|
|
 |
92.4 |
29 |
PDB header:antitoxin
Chain: B: PDB Molecule:antitoxin hicb;
PDBTitle: crystal structure of the burkholderia pseudomallei antitoxin hicb
|
|
|
|
| 11 | c2k5jB_
|
|
 |
88.4 |
23 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein yiif;
PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
|
|
|
|
| 12 | c4me7E_
|
|
 |
82.1 |
31 |
PDB header:hydrolase/hydrolase inhibitor
Chain: E: PDB Molecule:antitoxin endoai;
PDBTitle: crystal structure of bacillus subtilis toxin mazf in complex with2 cognate antitoxin maze
|
|
|
|
| 13 | c4p7dA_
|
|
 |
78.0 |
25 |
PDB header:toxin
Chain: A: PDB Molecule:antitoxin hicb3;
PDBTitle: antitoxin hicb3 crystal structure
|
|
|
|
| 14 | c6a6xC_
|
|
 |
55.0 |
30 |
PDB header:toxin
Chain: C: PDB Molecule:antitoxin maze7;
PDBTitle: the crystal structure of the mtb maze-mazf-mt9 complex
|
|
|
|
| 15 | d2cpga_
|
|
 |
51.9 |
30 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
|
|
|
| 16 | c1ea4K_
|
|
 |
51.9 |
30 |
PDB header:gene regulation/dna
Chain: K: PDB Molecule:transcriptional repressor copg;
PDBTitle: transcriptional repressor copg/22bp dsdna complex
|
|
|
|
| 17 | c6qeqD_
|
|
 |
51.6 |
14 |
PDB header:dna binding protein
Chain: D: PDB Molecule:pcff;
PDBTitle: pcff from enterococcus faecalis pcf10
|
|
|
|
| 18 | c2mdvB_
|
|
 |
27.1 |
33 |
PDB header:de novo protein
Chain: B: PDB Molecule:designed protein;
PDBTitle: nmr structure of beta alpha alpha 38
|
|
|
|
| 19 | d2vy4a1
|
|
 |
21.2 |
27 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:CHHC finger |
|
|
|
| 20 | c5cwwC_
|
|
 |
16.0 |
33 |
PDB header:transport protein
Chain: C: PDB Molecule:nucleoporin nup159;
PDBTitle: crystal structure of the chaetomium thermophilum heterotrimeric nup822 ntd-nup159 tail-nup145n apd complex
|
|
|
|
| 21 | d1jdfa2 |
|
not modelled |
15.0 |
27 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 22 | c1mszA_ |
|
not modelled |
14.9 |
21 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna-binding protein smubp-2;
PDBTitle: solution structure of the r3h domain from human smubp-2
|
|
|
| 23 | d1msza_ |
|
not modelled |
14.9 |
21 |
Fold:IF3-like
Superfamily:R3H domain
Family:R3H domain |
|
|
| 24 | d1rvka2 |
|
not modelled |
13.3 |
33 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 25 | c4l6tA_ |
|
not modelled |
13.0 |
46 |
PDB header:hydrolase
Chain: A: PDB Molecule:ecxa;
PDBTitle: gm1 bound form of the ecx ab5 holotoxin
|
|
|
| 26 | d2chra2 |
|
not modelled |
12.9 |
27 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 27 | d2hh6a1 |
|
not modelled |
12.6 |
32 |
Fold:Left-handed superhelix
Superfamily:BH3980-like
Family:BH3980-like |
|
|
| 28 | d1t98a1 |
|
not modelled |
12.5 |
35 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MukF N-terminal domain-like |
|
|
| 29 | d1bqga2 |
|
not modelled |
12.0 |
27 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 30 | d1jpma2 |
|
not modelled |
10.6 |
33 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 31 | d1wuea2 |
|
not modelled |
10.3 |
40 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 32 | c1t98B_ |
|
not modelled |
10.2 |
33 |
PDB header:cell cycle
Chain: B: PDB Molecule:chromosome partition protein mukf;
PDBTitle: crystal structure of mukf(1-287)
|
|
|
| 33 | c2rrfA_ |
|
not modelled |
10.2 |
14 |
PDB header:unknown function
Chain: A: PDB Molecule:zinc finger fyve domain-containing protein 21;
PDBTitle: the solution structure of the c-terminal region of zinc finger fyve2 domain-containing protein 21
|
|
|
| 34 | d2gl5a2 |
|
not modelled |
9.9 |
40 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 35 | d2mnra2 |
|
not modelled |
9.9 |
40 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 36 | d1nu5a2 |
|
not modelled |
9.8 |
27 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 37 | d1wufa2 |
|
not modelled |
9.7 |
27 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 38 | d2b0la1 |
|
not modelled |
9.6 |
47 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:CodY HTH domain |
|
|
| 39 | d1lqaa_ |
|
not modelled |
8.8 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
|
|
| 40 | d1yeya2 |
|
not modelled |
8.8 |
27 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 41 | c4n2sA_ |
|
not modelled |
8.6 |
18 |
PDB header:splicing/rna
Chain: A: PDB Molecule:tha8 rna binding protein;
PDBTitle: crystal structure of tha8 in complex with zm1a-6 rna
|
|
|
| 42 | d1sjda2 |
|
not modelled |
8.3 |
27 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 43 | c5b0nA_ |
|
not modelled |
8.1 |
26 |
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase ipah9.8;
PDBTitle: structure of shigella effector lrr domain
|
|
|
| 44 | d2c5ra1 |
|
not modelled |
7.9 |
50 |
Fold:Phage replication organizer domain
Superfamily:Phage replication organizer domain
Family:Phage replication organizer domain |
|
|
| 45 | c3jviA_ |
|
not modelled |
7.8 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein tyrosine phosphatase;
PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
|
|
|
| 46 | c4h3uB_ |
|
not modelled |
7.5 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of hypothetical protein with ketosteroid isomerase-2 like protein fold from catenulispora acidiphila dsm 44928
|
|
|
| 47 | d2gdqa2 |
|
not modelled |
6.9 |
20 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 48 | d1jpdx2 |
|
not modelled |
6.7 |
27 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 49 | d2ezwa1 |
|
not modelled |
6.5 |
28 |
Fold:Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
Superfamily:Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
Family:Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit |
|
|
| 50 | c4f9kA_ |
|
not modelled |
6.4 |
22 |
PDB header:transferase regulator
Chain: A: PDB Molecule:camp-dependent protein kinase type i-beta regulatory
PDBTitle: crystal structure of human camp-dependent protein kinase type i-beta2 regulatory subunit (fragment 11-73), northeast structural genomics3 consortium (nesg) target hr8613a
|
|
|
| 51 | c5ey0A_ |
|
not modelled |
6.4 |
35 |
PDB header:transcription
Chain: A: PDB Molecule:gtp-sensing transcriptional pleiotropic repressor cody;
PDBTitle: crystal structure of cody from staphylococcus aureus with gtp and ile
|
|
|
| 52 | d1muca2 |
|
not modelled |
6.3 |
60 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 53 | c3tadB_ |
|
not modelled |
6.1 |
20 |
PDB header:protein binding
Chain: B: PDB Molecule:liprin-alpha-2;
PDBTitle: crystal structure of the liprin-alpha/liprin-beta complex
|
|
|
| 54 | d1tzza2 |
|
not modelled |
6.1 |
53 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 55 | d1r0ma2 |
|
not modelled |
6.0 |
27 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 56 | c4zriC_ |
|
not modelled |
5.8 |
50 |
PDB header:signaling protein/transferase
Chain: C: PDB Molecule:serine/threonine-protein kinase lats2;
PDBTitle: crystal structure of merlin-ferm and lats2
|
|
|
| 57 | c4zriD_ |
|
not modelled |
5.7 |
50 |
PDB header:signaling protein/transferase
Chain: D: PDB Molecule:serine/threonine-protein kinase lats2;
PDBTitle: crystal structure of merlin-ferm and lats2
|
|
|
| 58 | c5o5jR_ |
|
not modelled |
5.7 |
30 |
PDB header:ribosome
Chain: R: PDB Molecule:30s ribosomal protein s18 2;
PDBTitle: structure of the 30s small ribosomal subunit from mycobacterium2 smegmatis
|
|
|
| 59 | d1h99a1 |
|
not modelled |
5.6 |
30 |
Fold:PTS-regulatory domain, PRD
Superfamily:PTS-regulatory domain, PRD
Family:PTS-regulatory domain, PRD |
|
|
| 60 | d2fq4a2 |
|
not modelled |
5.6 |
9 |
Fold:Tetracyclin repressor-like, C-terminal domain
Superfamily:Tetracyclin repressor-like, C-terminal domain
Family:Tetracyclin repressor-like, C-terminal domain |
|
|
| 61 | c2pijB_ |
|
not modelled |
5.3 |
24 |
PDB header:transcription
Chain: B: PDB Molecule:prophage pfl 6 cro;
PDBTitle: structure of the cro protein from prophage pfl 6 in pseudomonas2 fluorescens pf-5
|
|
|
| 62 | c3h87D_ |
|
not modelled |
5.3 |
39 |
PDB header:toxin/antitoxin
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: rv0301 rv0300 toxin antitoxin complex from mycobacterium tuberculosis
|
|
|
| 63 | c6g1oA_ |
|
not modelled |
5.3 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:isocitrate lyase;
PDBTitle: structure of pseudomonas aeruginosa isocitrate lyase, icl
|
|
|
| 64 | d2ftra1 |
|
not modelled |
5.2 |
27 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:EthD-like |
|
|
| 65 | d1q2za_ |
|
not modelled |
5.1 |
20 |
Fold:alpha-alpha superhelix
Superfamily:C-terminal domain of Ku80
Family:C-terminal domain of Ku80 |
|
|