1 c6ig0H_
100.0
30
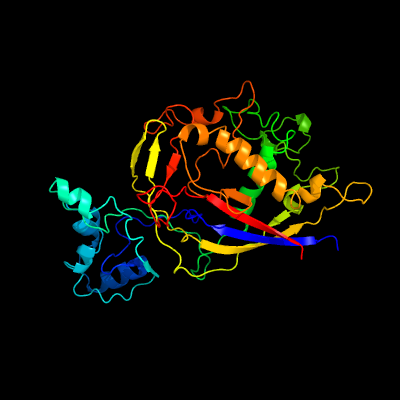
PDB header: rna binding proteinChain: H: PDB Molecule: type iii-a crispr-associated ramp protein csm5;PDBTitle: type iii-a csm complex, cryo-em structure of csm-ctr1, atp bound
2 c6musF_
100.0
22
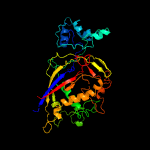
PDB header: rna binding protein/rnaChain: F: PDB Molecule: uncharacterized protein csm5;PDBTitle: cryo-em structure of larger csm-crrna-target rna ternary complex in2 type iii-a crispr-cas system
3 c6musK_
99.6
21
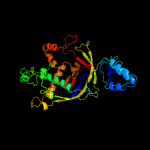
PDB header: rna binding protein/rnaChain: K: PDB Molecule: uncharacterized protein csm3;PDBTitle: cryo-em structure of larger csm-crrna-target rna ternary complex in2 type iii-a crispr-cas system
4 c4w8vA_
99.6
18
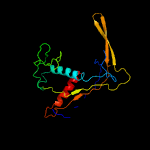
PDB header: rna binding proteinChain: A: PDB Molecule: crispr system cmr subunit cmr6;PDBTitle: crystal structure of cmr6 from pyrococcus furiosus
5 c6ifnF_
99.5
22
PDB header: rna binding proteinChain: F: PDB Molecule: type iii-a crispr-associated ramp protein csm3;PDBTitle: crystal structure of type iii-a crispr csm complex
6 c4qtsC_
99.4
23
PDB header: rna binding proteinChain: C: PDB Molecule: crispr type iii-associated ramp protein csm3;PDBTitle: crystal structure of csm3-csm4 subcomplex in the type iii-a crispr-cas2 interference complex
7 c5yjdB_
99.4
20
PDB header: hydrolaseChain: B: PDB Molecule: csm3;PDBTitle: structural insights into the crispr-cas-associated ribonuclease2 activity of staphylococcus epidermidis csm3
8 c6ae2B_
99.3
16
PDB header: rna binding proteinChain: B: PDB Molecule: csm3;PDBTitle: crystal structure of csm3 of the type iii-a crispr-cas effector2 complex
9 c3x1lE_
99.3
27
PDB header: rna binding protein/rna/dnaChain: E: PDB Molecule: cmr4;PDBTitle: crystal structure of the crispr-cas rna silencing cmr complex bound to2 a target analog
10 c4n0lB_
99.0
18
PDB header: rna binding proteinChain: B: PDB Molecule: predicted component of a thermophile-specific dna repairPDBTitle: methanopyrus kandleri csm3 crystal structure
11 c4w8wB_
99.0
39
PDB header: hydrolaseChain: B: PDB Molecule: crispr system cmr subunit cmr4;PDBTitle: crystal structure of oligomeric cmr4 from pyrococcus furiosus
12 c3x1lH_
98.9
13
PDB header: rna binding protein/rna/dnaChain: H: PDB Molecule: cmr6;PDBTitle: crystal structure of the crispr-cas rna silencing cmr complex bound to2 a target analog
13 c4w8xA_
83.4
5
PDB header: rna binding proteinChain: A: PDB Molecule: crispr system cmr subunit cmr1-1;PDBTitle: crystal structure of cmr1 from pyrococcus furiosus bound to a2 nucleotide
14 c3ijeB_
77.3
15
PDB header: protein bindingChain: B: PDB Molecule: integrin beta-3;PDBTitle: crystal structure of the complete integrin alhavbeta3 ectodomain plus2 an alpha/beta transmembrane fragment
15 c3k6sB_
60.2
21
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-2;PDBTitle: structure of integrin alphaxbeta2 ectodomain
16 c1u3fA_
58.3
38
PDB header: ligaseChain: A: PDB Molecule: 5,10-methenyltetrahydrofolate synthetase;PDBTitle: structural and functional characterization of a 5,10-2 methenyltetrahydrofolate synthetase from mycoplasma3 pneumoniae (gi: 13508087)
17 d1sbqa_
58.3
38
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: Methenyltetrahydrofolate synthetase
18 c2jcbA_
52.2
31
PDB header: ligaseChain: A: PDB Molecule: 5-formyltetrahydrofolate cyclo-ligase family protein;PDBTitle: the crystal structure of 5-formyl-tetrahydrofolate2 cycloligase from bacillus anthracis (ba4489)
19 c3hy4A_
50.1
38
PDB header: ligaseChain: A: PDB Molecule: 5-formyltetrahydrofolate cyclo-ligase;PDBTitle: structure of human mthfs with n5-iminium phosphate
20 d1wkca_
48.7
31
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: Methenyltetrahydrofolate synthetase
21 c1ydmC_
not modelled
45.7
31
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein yqgn;PDBTitle: x-ray structure of northeast structural genomics target sr44
22 c1u8cB_
not modelled
40.9
15
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: a novel adaptation of the integrin psi domain revealed from its2 crystal structure
23 c2q6mA_
not modelled
39.8
30
PDB header: toxinChain: A: PDB Molecule: cholix toxin;PDBTitle: catalytic fragment of cholix toxin from vibrio cholerae in complex2 with the pj34 inhibitor
24 d1soua_
not modelled
38.8
38
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: Methenyltetrahydrofolate synthetase
25 c4l6uB_
not modelled
36.9
33
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of af1868: cmr1 subunit of the cmr rna silencing2 complex
26 c4h4kA_
not modelled
33.5
29
PDB header: rna binding proteinChain: A: PDB Molecule: crispr system cmr subunit cmr3;PDBTitle: structure of the cmr2-cmr3 subcomplex of the cmr rna-silencing complex
27 d1qr0a1
not modelled
30.7
31
Fold: 4'-phosphopantetheinyl transferaseSuperfamily: 4'-phosphopantetheinyl transferaseFamily: 4'-Phosphopantetheinyl transferase SFP
28 c2q5tA_
not modelled
30.7
30
PDB header: toxinChain: A: PDB Molecule: cholix toxin;PDBTitle: full-length cholix toxin from vibrio cholerae
29 c2lycA_
not modelled
28.6
67
PDB header: protein bindingChain: A: PDB Molecule: spindle and kinetochore-associated protein 1 homolog;PDBTitle: structure of c-terminal domain of ska1
30 d1ikpa2
not modelled
25.3
41
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins
31 d2ix0a3
not modelled
22.0
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
32 c4um9D_
not modelled
20.7
22
PDB header: immune systemChain: D: PDB Molecule: integrin beta-6;PDBTitle: crystal structure of alpha v beta 6 with peptide
33 c6n1nA_
not modelled
19.1
30
PDB header: hydrolaseChain: A: PDB Molecule: beta-lactamase;PDBTitle: crystal structure of class d beta-lactamase from sebaldella termitidis2 atcc 33386
34 c2it0A_
not modelled
18.7
42
PDB header: transcription/dnaChain: A: PDB Molecule: iron-dependent repressor ider;PDBTitle: crystal structure of a two-domain ider-dna complex crystal2 form ii
35 c5bv3C_
not modelled
18.0
13
PDB header: hydrolaseChain: C: PDB Molecule: m7gpppx diphosphatase;PDBTitle: yeast scavenger decapping enzyme in complex with m7gdp
36 c1ikqA_
not modelled
16.9
41
PDB header: transferaseChain: A: PDB Molecule: exotoxin a;PDBTitle: pseudomonas aeruginosa exotoxin a, wild type
37 c1qr0A_
not modelled
16.3
29
PDB header: transferaseChain: A: PDB Molecule: 4'-phosphopantetheinyl transferase sfp;PDBTitle: crystal structure of the 4'-phosphopantetheinyl transferase sfp-2 coenzyme a complex
38 c2ixsB_
not modelled
15.9
25
PDB header: hydrolaseChain: B: PDB Molecule: sdai restriction endonuclease;PDBTitle: structure of sdai restriction endonuclease
39 c1cjyB_
not modelled
15.9
11
PDB header: hydrolaseChain: B: PDB Molecule: protein (cytosolic phospholipase a2);PDBTitle: human cytosolic phospholipase a2
40 c4gn2A_
not modelled
15.8
27
PDB header: hydrolaseChain: A: PDB Molecule: oxacillinase;PDBTitle: crystal structure of oxa-45, a class d beta-lactamase with extended2 spectrum activity
41 c4xpuA_
not modelled
14.9
80
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease v;PDBTitle: the crystal structure of endov from e.coli
42 c3ga2A_
not modelled
14.6
80
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease v;PDBTitle: crystal structure of the endonuclease_v (bsu36170) from bacillus2 subtilis, northeast structural genomics consortium target sr624
43 c2mmbA_
not modelled
14.4
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of the protein yp_001712342.1 from acinetobacter2 baumannii
44 c4xb6D_
not modelled
14.3
36
PDB header: transferaseChain: D: PDB Molecule: alpha-d-ribose 1-methylphosphonate 5-phosphate c-p lyase;PDBTitle: structure of the e. coli c-p lyase core complex
45 d1rutx3
not modelled
14.2
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain
46 c3jszA_
not modelled
14.2
12
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: legionella pneumophila glucosyltransferase lgt1 n293a with udp-glc
47 c4nspA_
not modelled
14.1
80
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease v;PDBTitle: crystal structure of human endov
48 c2moqA_
not modelled
13.6
11
PDB header: protein bindingChain: A: PDB Molecule: iron-regulated surface determinant protein b;PDBTitle: solution structure and molecular determinants of hemoglobin binding of2 the first neat domain of isdb in staphylococcus aureus
49 d1k38a_
not modelled
13.5
40
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase
50 d1isua_
not modelled
13.4
50
Fold: HIPIP (high potential iron protein)Superfamily: HIPIP (high potential iron protein)Family: HIPIP (high potential iron protein)
51 c4khbF_
not modelled
13.3
63
PDB header: transcription/replicationChain: F: PDB Molecule: uncharacterized protein pob3n;PDBTitle: structure of the spt16d pob3n heterodimer
52 c6bn3A_
not modelled
13.2
29
PDB header: hydrolaseChain: A: PDB Molecule: beta-lactamase;PDBTitle: ctx-m-151 class a extended-spectrum beta-lactamase apo crystal2 structure at 1.3 angstrom resolution.
53 c3gocB_
not modelled
13.1
100
PDB header: hydrolaseChain: B: PDB Molecule: endonuclease v;PDBTitle: crystal structure of the endonuclease v (sav1684) from streptomyces2 avermitilis. northeast structural genomics consortium target svr196
54 c2w36B_
not modelled
13.0
80
PDB header: hydrolaseChain: B: PDB Molecule: endonuclease v;PDBTitle: structures of endonuclease v with dna reveal initiation of2 deaminated adenine repair
55 c5u1lA_
not modelled
12.8
10
PDB header: membrane proteinChain: A: PDB Molecule: p2x purinoceptor;PDBTitle: crystal structure of the atp-gated p2x7 ion channel in the closed, apo2 state
56 c4jbfB_
not modelled
12.7
20
PDB header: transferaseChain: B: PDB Molecule: peptidoglycan glycosyltransferase;PDBTitle: crystal structure of peptidoglycan glycosyltransferase from atopobium2 parvulum dsm 20469.
57 c1cm4B_
not modelled
12.6
30
PDB header: calcium-binding/transferaseChain: B: PDB Molecule: calmodulin-dependent protein kinase ii-alpha;PDBTitle: motions of calmodulin-four-conformer refinement
58 c1cm1B_
not modelled
12.6
30
PDB header: complex (calcium-binding/transferase)Chain: B: PDB Molecule: calmodulin-dependent protein kinase ii-alpha;PDBTitle: motions of calmodulin-single-conformer refinement
59 c1cdmB_
not modelled
12.6
30
PDB header: calcium-binding proteinChain: B: PDB Molecule: calmodulin;PDBTitle: modulation of calmodulin plasticity in molecular2 recognition on the basis of x-ray structures
60 c4k0xA_
not modelled
12.5
47
PDB header: hydrolaseChain: A: PDB Molecule: beta-lactamase;PDBTitle: x-ray crystal structure of oxa-23 from acinetobacter baumannii
61 c3gp2B_
not modelled
12.5
30
PDB header: metal binding protein/transferaseChain: B: PDB Molecule: calcium/calmodulin-dependent protein kinase typePDBTitle: calmodulin bound to peptide from calmodulin kinase ii2 (camkii)
62 c5xw6B_
not modelled
12.5
14
PDB header: membrane proteinChain: B: PDB Molecule: p2x purinoceptor;PDBTitle: crystal structure of the chicken atp-gated p2x7 receptor channel in2 the presence of competitive antagonist tnp-atp at 3.1 angstroms
63 d1r5ba3
not modelled
12.3
40
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins
64 c4hlxD_
not modelled
12.2
12
PDB header: dna binding proteinChain: D: PDB Molecule: k9;PDBTitle: the crystal structure of the dna binding domain of virf-1 from the2 oncogenic kshv
65 d1xa1a_
not modelled
12.1
37
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase
66 c3i5dC_
not modelled
11.9
14
PDB header: transport proteinChain: C: PDB Molecule: p2x purinoceptor;PDBTitle: crystal structure of the atp-gated p2x4 ion channel in the closed, apo2 state at 3.5 angstroms (r3)
67 c6gnyD_
not modelled
11.6
33
PDB header: structural proteinChain: D: PDB Molecule: telomere repeats-binding bouquet formation protein 2;PDBTitle: crystal structure of the majin-terb2 heterotetrameric complex
68 c2wd6B_
not modelled
11.6
24
PDB header: cell adhesionChain: B: PDB Molecule: agglutinin receptor;PDBTitle: crystal structure of the variable domain of the streptococcus gordonii2 surface protein sspb
69 c6gnxB_
not modelled
11.5
33
PDB header: structural proteinChain: B: PDB Molecule: telomere repeats-binding bouquet formation protein 2;PDBTitle: crystal structure of the majin-terb2 heterotetrameric complex -2 selenomethionine derivative
70 c3fcuB_
not modelled
11.3
15
PDB header: cell adhesion/blood clottingChain: B: PDB Molecule: integrin beta-3;PDBTitle: structure of headpiece of integrin aiibb3 in open conformation
71 c3vi3D_
not modelled
10.6
14
PDB header: cell adhesion/immune systemChain: D: PDB Molecule: integrin beta-1;PDBTitle: crystal structure of alpha5beta1 integrin headpiece (ligand-free form)
72 d2pifa1
not modelled
10.1
36
Fold: PSTPO5379-likeSuperfamily: PSTPO5379-likeFamily: PSTPO5379-like
73 c4kt3B_
not modelled
9.8
42
PDB header: hydrolaseChain: B: PDB Molecule: putative lipoprotein;PDBTitle: structure of a type vi secretion system effector-immunity complex from2 pseudomonas protegens
74 c3oqcB_
not modelled
9.6
63
PDB header: hydrolaseChain: B: PDB Molecule: ufm1-specific protease 2;PDBTitle: ubiquitin-fold modifier 1 specific protease, ufsp2
75 c6nhsA_
not modelled
9.6
22
PDB header: hydrolaseChain: A: PDB Molecule: beta-lactamase;PDBTitle: crystal structure of the beta lactamase class d ybxi from nostoc
76 c4oh0A_
not modelled
9.6
40
PDB header: hydrolaseChain: A: PDB Molecule: beta-lactamase oxa-58;PDBTitle: crystal structure of oxa-58 carbapenemase
77 c2jc7A_
not modelled
9.5
44
PDB header: hydrolaseChain: A: PDB Molecule: beta-lactamase oxa-24;PDBTitle: the crystal structure of the carbapenemase oxa-24 reveals2 new insights into the mechanism of carbapenem-hydrolysis
78 c3j7yf_
not modelled
9.0
21
PDB header: ribosomeChain: F: PDB Molecule: ul4;PDBTitle: structure of the large ribosomal subunit from human mitochondria
79 c4byrP_
not modelled
8.9
50
PDB header: ribosomeChain: P: PDB Molecule: eukaryotic translation initiation factor 5b;PDBTitle: cryo-em reconstruction of the 80s-eif5b-met-itrnamet2 eukaryotic translation initiation complex
80 d1itza3
not modelled
8.8
30
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Transketolase C-terminal domain-like
81 c5ctmB_
not modelled
8.7
30
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase;PDBTitle: structure of bpu1 beta-lactamase
82 d1irxa2
not modelled
8.6
25
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain
83 c2hdnJ_
not modelled
8.6
50
PDB header: translationChain: J: PDB Molecule: elongation factor ef-tu;PDBTitle: trypsin-modified elongation factor tu in complex with2 tetracycline at 2.8 angstrom resolution
84 c2z84A_
not modelled
8.5
63
PDB header: hydrolaseChain: A: PDB Molecule: ufm1-specific protease 1;PDBTitle: insights from crystal and solution structures of mouse ufsp1
85 d1rr7a_
not modelled
8.3
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Middle operon regulator, Mor
86 c1rr7A_
not modelled
8.3
21
PDB header: transcriptionChain: A: PDB Molecule: middle operon regulator;PDBTitle: crystal structure of the middle operon regulator protein of2 bacteriophage mu
87 c2xi5D_
not modelled
8.1
24
PDB header: transferaseChain: D: PDB Molecule: rna polymerase l;PDBTitle: n-terminal endonuclease domain of la crosse virus l-protein
88 c5cv1A_
not modelled
8.1
20
PDB header: hydrolaseChain: A: PDB Molecule: p granule abnormality protein 1;PDBTitle: c. elegans pgl-1 dimerization domain
89 d1tuba1
not modelled
8.0
29
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain
90 c2xi7B_
not modelled
8.0
24
PDB header: transferaseChain: B: PDB Molecule: rna polymerase l;PDBTitle: n-terminal endonuclease domain of la crosse virus l-protein
91 c3c26A_
not modelled
8.0
50
PDB header: transferaseChain: A: PDB Molecule: putative acetyltransferase ta0821;PDBTitle: crystal structure of a putative acetyltransferase (np_394282.1) from2 thermoplasma acidophilum at 2.00 a resolution
92 c6k0bH_
not modelled
8.0
22
PDB header: rna binding protein/rnaChain: H: PDB Molecule: ribonuclease p protein component 4;PDBTitle: cryo-em structure of archaeal ribonuclease p with mature trna
93 c5k0yS_
not modelled
7.8
25
PDB header: translationChain: S: PDB Molecule: eukaryotic initiation factor 2 gamma subunit (eif2-gamma);PDBTitle: m48s late-stage initiation complex, purified from rabbit reticulocytes2 lysates, displaying eif2 ternary complex and eif3 i and g subunits3 relocated to the intersubunit face
94 c6ei1A_
not modelled
7.8
63
PDB header: hydrolaseChain: A: PDB Molecule: zinc finger with ufm1-specific peptidase domain protein;PDBTitle: crystal structure of the covalent complex between deubiquitinase zufsp2 (zup1) and ubiquitin-pa
95 c3hbrD_
not modelled
7.7
30
PDB header: hydrolaseChain: D: PDB Molecule: oxa-48;PDBTitle: crystal structure of oxa-48 beta-lactamase
96 c6gbsB_
not modelled
7.7
12
PDB header: hydrolaseChain: B: PDB Molecule: putative mrna decapping protein;PDBTitle: crystal structure of the c. themophilum scavenger decapping enzyme2 dcps apo form
97 c2cg5A_
not modelled
7.6
14
PDB header: transferase/hydrolaseChain: A: PDB Molecule: l-aminoadipate-semialdehyde dehydrogenase-PDBTitle: structure of aminoadipate-semialdehyde dehydrogenase-2 phosphopantetheinyl transferase in complex with cytosolic acyl3 carrier protein and coenzyme a
98 d1tubb1
not modelled
7.5
18
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain
99 c5e2fB_
not modelled
7.4
17
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase ybxi;PDBTitle: crystal structure of beta-lactamase class d from bacillus subtilis