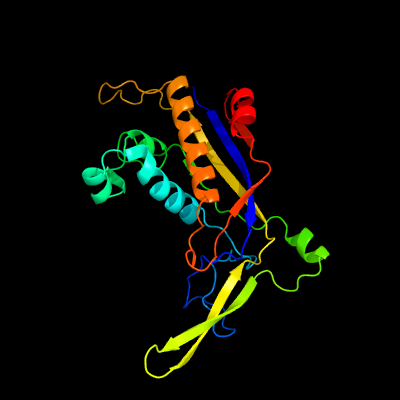
| 1 | c6ifnF_
|
|
 |
100.0 |
50 |
PDB header:rna binding protein
Chain: F: PDB Molecule:type iii-a crispr-associated ramp protein csm3;
PDBTitle: crystal structure of type iii-a crispr csm complex
|
|
|
|
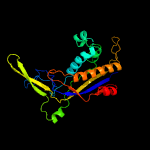
| 2 | c4n0lB_
|
|
 |
100.0 |
30 |
PDB header:rna binding protein
Chain: B: PDB Molecule:predicted component of a thermophile-specific dna repair
PDBTitle: methanopyrus kandleri csm3 crystal structure
|
|
|
|
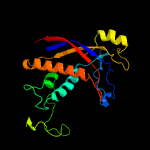
| 3 | c6musK_
|
|
 |
100.0 |
40 |
PDB header:rna binding protein/rna
Chain: K: PDB Molecule:uncharacterized protein csm3;
PDBTitle: cryo-em structure of larger csm-crrna-target rna ternary complex in2 type iii-a crispr-cas system
|
|
|
|
| 4 | c5yjdB_
|
|
 |
100.0 |
40 |
PDB header:hydrolase
Chain: B: PDB Molecule:csm3;
PDBTitle: structural insights into the crispr-cas-associated ribonuclease2 activity of staphylococcus epidermidis csm3
|
|
|
|
| 5 | c4qtsC_
|
|
 |
100.0 |
38 |
PDB header:rna binding protein
Chain: C: PDB Molecule:crispr type iii-associated ramp protein csm3;
PDBTitle: crystal structure of csm3-csm4 subcomplex in the type iii-a crispr-cas2 interference complex
|
|
|
|
| 6 | c6ae2B_
|
|
 |
100.0 |
32 |
PDB header:rna binding protein
Chain: B: PDB Molecule:csm3;
PDBTitle: crystal structure of csm3 of the type iii-a crispr-cas effector2 complex
|
|
|
|
| 7 | c4w8vA_
|
|
 |
100.0 |
21 |
PDB header:rna binding protein
Chain: A: PDB Molecule:crispr system cmr subunit cmr6;
PDBTitle: crystal structure of cmr6 from pyrococcus furiosus
|
|
|
|
| 8 | c3x1lE_
|
|
 |
100.0 |
27 |
PDB header:rna binding protein/rna/dna
Chain: E: PDB Molecule:cmr4;
PDBTitle: crystal structure of the crispr-cas rna silencing cmr complex bound to2 a target analog
|
|
|
|
| 9 | c4w8wB_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:crispr system cmr subunit cmr4;
PDBTitle: crystal structure of oligomeric cmr4 from pyrococcus furiosus
|
|
|
|
| 10 | c3x1lH_
|
|
 |
99.9 |
24 |
PDB header:rna binding protein/rna/dna
Chain: H: PDB Molecule:cmr6;
PDBTitle: crystal structure of the crispr-cas rna silencing cmr complex bound to2 a target analog
|
|
|
|
| 11 | c6ig0H_
|
|
 |
99.7 |
24 |
PDB header:rna binding protein
Chain: H: PDB Molecule:type iii-a crispr-associated ramp protein csm5;
PDBTitle: type iii-a csm complex, cryo-em structure of csm-ctr1, atp bound
|
|
|
|
| 12 | c4w8xA_
|
|
 |
99.6 |
18 |
PDB header:rna binding protein
Chain: A: PDB Molecule:crispr system cmr subunit cmr1-1;
PDBTitle: crystal structure of cmr1 from pyrococcus furiosus bound to a2 nucleotide
|
|
|
|
| 13 | c6musF_
|
|
 |
99.5 |
26 |
PDB header:rna binding protein/rna
Chain: F: PDB Molecule:uncharacterized protein csm5;
PDBTitle: cryo-em structure of larger csm-crrna-target rna ternary complex in2 type iii-a crispr-cas system
|
|
|
|
| 14 | c4l6uB_
|
|
 |
99.5 |
21 |
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of af1868: cmr1 subunit of the cmr rna silencing2 complex
|
|
|
|
| 15 | c6muuE_
|
|
 |
98.4 |
19 |
PDB header:rna binding protein/rna
Chain: E: PDB Molecule:uncharacterized protein csm4;
PDBTitle: cryo-em structure of csm-crrna binary complex in type iii-a crispr-cas2 system
|
|
|
|
| 16 | c6ig0B_
|
|
 |
97.9 |
18 |
PDB header:rna binding protein
Chain: B: PDB Molecule:type iii-a crispr-associated ramp protein csm4;
PDBTitle: type iii-a csm complex, cryo-em structure of csm-ctr1, atp bound
|
|
|
|
| 17 | c4qtsB_
|
|
 |
97.7 |
24 |
PDB header:rna binding protein
Chain: B: PDB Molecule:crispr type iii-associated ramp protein csm4;
PDBTitle: crystal structure of csm3-csm4 subcomplex in the type iii-a crispr-cas2 interference complex
|
|
|
|
| 18 | c6fjwA_
|
|
 |
94.7 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:cas6 protein;
PDBTitle: streptococcus thermophilus cas6
|
|
|
|
| 19 | c4c97B_
|
|
 |
92.1 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:cas6a;
PDBTitle: cas6 (ttha0078) h37a mutant
|
|
|
|
| 20 | c4ilmH_
|
|
 |
92.1 |
16 |
PDB header:hydrolase/rna
Chain: H: PDB Molecule:crispr-associated endoribonuclease cas6 2;
PDBTitle: crispr rna processing endoribonuclease
|
|
|
|
| 21 | c6njyB_ |
|
not modelled |
87.1 |
15 |
PDB header:rna binding protein
Chain: B: PDB Molecule:type iv crispr associated cas6 rna endonuclease;
PDBTitle: type iv crispr associated rna endonuclease cas6 - apo form
|
|
|
| 22 | c3i4hX_ |
|
not modelled |
79.6 |
27 |
PDB header:hydrolase
Chain: X: PDB Molecule:endoribonuclease;
PDBTitle: crystal structure of cas6 in pyrococcus furiosus
|
|
|
| 23 | c3qjjB_ |
|
not modelled |
77.3 |
19 |
PDB header:immune system/rna
Chain: B: PDB Molecule:putative uncharacterized protein ph0350;
PDBTitle: one ramp protein binding different rna substrates
|
|
|
| 24 | c4txdA_ |
|
not modelled |
76.7 |
23 |
PDB header:rna binding protein
Chain: A: PDB Molecule:csc2;
PDBTitle: crystal structure of thermofilum pendens csc2
|
|
|
| 25 | c4c9dA_ |
|
not modelled |
74.1 |
23 |
PDB header:hydrolase/rna
Chain: A: PDB Molecule:cas6b;
PDBTitle: cas6 (tthb231) product complex
|
|
|
| 26 | c1ajnA_ |
|
not modelled |
74.0 |
17 |
PDB header:antibiotic resistance
Chain: A: PDB Molecule:penicillin amidohydrolase;
PDBTitle: penicillin acylase complexed with p-nitrophenylacetic acid
|
|
|
| 27 | c5yi6A_ |
|
not modelled |
72.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:crispr-associated endoribonuclease cas6 1;
PDBTitle: crispr associated protein cas6
|
|
|
| 28 | c1cp9A_ |
|
not modelled |
71.1 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:penicillin g amidase;
PDBTitle: crystal structure of penicillin g acylase from the bro1 mutant strain2 of providencia rettgeri
|
|
|
| 29 | c4z7lB_ |
|
not modelled |
71.0 |
33 |
PDB header:hydrolase/rna
Chain: B: PDB Molecule:cas6b;
PDBTitle: crystal structure of cas6b
|
|
|
| 30 | c1gk9A_ |
|
not modelled |
71.0 |
17 |
PDB header:antibiotic resistance
Chain: A: PDB Molecule:penicillin g acylase alpha subunit;
PDBTitle: crystal structures of penicillin acylase enzyme-substrate2 complexes: structural insights into the catalytic mechanism
|
|
|
| 31 | c1e3aA_ |
|
not modelled |
68.7 |
17 |
PDB header:antibiotic resistance
Chain: A: PDB Molecule:penicillin amidase alpha subunit;
PDBTitle: a slow processing precursor penicillin acylase from escherichia coli
|
|
|
| 32 | c4yfbD_ |
|
not modelled |
65.1 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:protein related to penicillin acylase;
PDBTitle: structure of n-acylhomoserine lactone acylase macq in complex with2 phenylacetic acid
|
|
|
| 33 | c6nvyC_ |
|
not modelled |
57.2 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:penicillin g acylase;
PDBTitle: crystal structure of penicillin g acylase from bacillus thermotolerans
|
|
|
| 34 | c2ae3A_ |
|
not modelled |
53.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:glutaryl 7-aminocephalosporanic acid acylase;
PDBTitle: glutaryl 7-aminocephalosporanic acid acylase: mutational study of2 activation mechanism
|
|
|
| 35 | c2wybA_ |
|
not modelled |
50.9 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:acyl-homoserine lactone acylase pvdq subunit
PDBTitle: the quorum quenching n-acyl homoserine lactone acylase pvdq2 with a covalently bound dodecanoic acid
|
|
|
| 36 | c3k3wA_ |
|
not modelled |
49.7 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:penicillin g acylase;
PDBTitle: thermostable penicillin g acylase from alcaligenes faecalis in2 orthorhombic form
|
|
|
| 37 | c4hsrA_ |
|
not modelled |
49.5 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:glutaryl-7-aminocephalosporanic acid acylase alpha chain;
PDBTitle: crystal structure of a class iii engineered cephalosporin acylase
|
|
|
| 38 | c1oqzB_ |
|
not modelled |
39.5 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:glutaryl acylase;
PDBTitle: crystal structures of glutaryl 7-aminocephalosporanic acid acylase:2 insight into autoproteolytic activation
|
|
|
| 39 | c6dd5B_ |
|
not modelled |
22.4 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:mmb-1 cas6 fused to maltose binding protein,crispr-
PDBTitle: crystal structure of the cas6 domain of marinomonas mediterranea mmb-12 cas6-rt-cas1 fusion protein
|
|
|
| 40 | c5il5A_ |
|
not modelled |
15.6 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:mlnd;
PDBTitle: crystal structure of the dehydratase domain of mlnd from bacillus2 amyloliquefaciens
|
|
|
| 41 | c4u3vA_ |
|
not modelled |
15.1 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:polyketide synthase pksr;
PDBTitle: crystal structure of the trans-acyltransferase polyketide synthase2 enoyl-isomerase
|
|
|
| 42 | c4uedB_ |
|
not modelled |
15.1 |
47 |
PDB header:translation
Chain: B: PDB Molecule:eukaryotic translation factor 4e-binding protein 1;
PDBTitle: complex of human eif4e with the 4e binding protein 4e-bp1
|
|
|
| 43 | c6h9iB_ |
|
not modelled |
14.4 |
45 |
PDB header:rna binding protein
Chain: B: PDB Molecule:csf5;
PDBTitle: csf5, crispr-cas type iv cas6 crrna endonuclease
|
|
|
| 44 | d1qr0a1 |
|
not modelled |
13.4 |
25 |
Fold:4'-phosphopantetheinyl transferase
Superfamily:4'-phosphopantetheinyl transferase
Family:4'-Phosphopantetheinyl transferase SFP |
|
|
| 45 | c5hu7B_ |
|
not modelled |
12.2 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:putative polyketide synthase;
PDBTitle: crystal structure of the trans-at pks dehydratase domain of c0zgq42 from brevibacillus brevis
|
|
|
| 46 | c3um0B_ |
|
not modelled |
10.6 |
50 |
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:charged multivesicular body protein 5;
PDBTitle: crystal structure of the brox bro1 domain in complex with the c-2 terminal tail of chmp5
|
|
|
| 47 | d2qamr1 |
|
not modelled |
10.4 |
28 |
Fold:L21p-like
Superfamily:L21p-like
Family:Ribosomal protein L21p |
|
|
| 48 | c3ga2A_ |
|
not modelled |
9.5 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease v;
PDBTitle: crystal structure of the endonuclease_v (bsu36170) from bacillus2 subtilis, northeast structural genomics consortium target sr624
|
|
|
| 49 | c3c26A_ |
|
not modelled |
9.3 |
45 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase ta0821;
PDBTitle: crystal structure of a putative acetyltransferase (np_394282.1) from2 thermoplasma acidophilum at 2.00 a resolution
|
|
|
| 50 | c4xpuA_ |
|
not modelled |
9.1 |
67 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease v;
PDBTitle: the crystal structure of endov from e.coli
|
|
|
| 51 | d1cixa_ |
|
not modelled |
8.6 |
54 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:omega toxin-like
Family:Spider toxins |
|
|
| 52 | c1cixA_ |
|
not modelled |
8.6 |
54 |
PDB header:antimicrobial peptide
Chain: A: PDB Molecule:protein (tachystatin a);
PDBTitle: three-dimensional structure of antimicrobial peptide2 tachystatin a isolated from horseshoe crab
|
|
|
| 53 | c4nspA_ |
|
not modelled |
8.5 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease v;
PDBTitle: crystal structure of human endov
|
|
|
| 54 | c3keyA_ |
|
not modelled |
8.4 |
23 |
PDB header:structural protein
Chain: A: PDB Molecule:protein stn1;
PDBTitle: crystal structure of s. cerevisiae stn1 c-terminal
|
|
|
| 55 | c6gnyD_ |
|
not modelled |
7.7 |
83 |
PDB header:structural protein
Chain: D: PDB Molecule:telomere repeats-binding bouquet formation protein 2;
PDBTitle: crystal structure of the majin-terb2 heterotetrameric complex
|
|
|
| 56 | c2w36B_ |
|
not modelled |
7.7 |
67 |
PDB header:hydrolase
Chain: B: PDB Molecule:endonuclease v;
PDBTitle: structures of endonuclease v with dna reveal initiation of2 deaminated adenine repair
|
|
|
| 57 | c6gnxB_ |
|
not modelled |
7.7 |
83 |
PDB header:structural protein
Chain: B: PDB Molecule:telomere repeats-binding bouquet formation protein 2;
PDBTitle: crystal structure of the majin-terb2 heterotetrameric complex -2 selenomethionine derivative
|
|
|
| 58 | d1r5ba3 |
|
not modelled |
7.6 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:G proteins |
|
|
| 59 | c3gocB_ |
|
not modelled |
7.5 |
50 |
PDB header:hydrolase
Chain: B: PDB Molecule:endonuclease v;
PDBTitle: crystal structure of the endonuclease v (sav1684) from streptomyces2 avermitilis. northeast structural genomics consortium target svr196
|
|
|
| 60 | c2lycA_ |
|
not modelled |
7.2 |
50 |
PDB header:protein binding
Chain: A: PDB Molecule:spindle and kinetochore-associated protein 1 homolog;
PDBTitle: structure of c-terminal domain of ska1
|
|
|
| 61 | c6qsrA_ |
|
not modelled |
6.8 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:amp-dependent synthetase and ligase;
PDBTitle: the dehydratase heterocomplex apei:p from xenorhabdus doucetiae
|
|
|
| 62 | c5mlcT_ |
|
not modelled |
6.8 |
10 |
PDB header:ribosome
Chain: T: PDB Molecule:50s ribosomal protein l21, chloroplastic;
PDBTitle: cryo-em structure of the spinach chloroplast ribosome reveals the2 location of plastid-specific ribosomal proteins and extensions
|
|
|
| 63 | c3bboT_ |
|
not modelled |
6.3 |
10 |
PDB header:ribosome
Chain: T: PDB Molecule:ribosomal protein l21;
PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
|
|
|
| 64 | d2ix0a3 |
|
not modelled |
6.2 |
43 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
|
|
| 65 | c4regA_ |
|
not modelled |
6.0 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure analysis of pf0642
|
|
|
| 66 | c5mmiS_ |
|
not modelled |
6.0 |
10 |
PDB header:ribosome
Chain: S: PDB Molecule:50s ribosomal protein l21, chloroplastic;
PDBTitle: structure of the large subunit of the chloroplast ribosome
|
|
|
| 67 | c4byrP_ |
|
not modelled |
5.7 |
11 |
PDB header:ribosome
Chain: P: PDB Molecule:eukaryotic translation initiation factor 5b;
PDBTitle: cryo-em reconstruction of the 80s-eif5b-met-itrnamet2 eukaryotic translation initiation complex
|
|
|
| 68 | c5bp2A_ |
|
not modelled |
5.6 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:mycocerosic acid synthase-like polyketide synthase;
PDBTitle: dehydratase domain (dh) of a mycocerosic acid synthase-like (mas-like)2 pks, crystal form 1
|
|
|