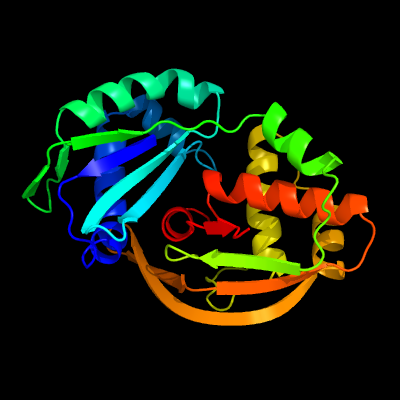
| 1 | c6fjwA_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:cas6 protein;
PDBTitle: streptococcus thermophilus cas6
|
|
|
|
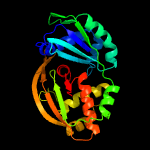
| 2 | c4c97B_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:cas6a;
PDBTitle: cas6 (ttha0078) h37a mutant
|
|
|
|
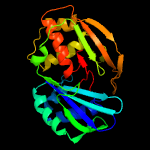
| 3 | c4c9dA_
|
|
 |
100.0 |
27 |
PDB header:hydrolase/rna
Chain: A: PDB Molecule:cas6b;
PDBTitle: cas6 (tthb231) product complex
|
|
|
|
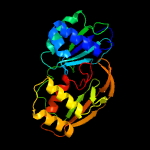
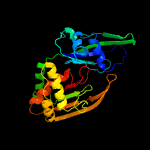
| 4 | c4ilmH_
|
|
 |
100.0 |
12 |
PDB header:hydrolase/rna
Chain: H: PDB Molecule:crispr-associated endoribonuclease cas6 2;
PDBTitle: crispr rna processing endoribonuclease
|
|
|
|
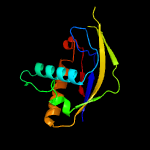
| 5 | c6njyB_
|
|
 |
100.0 |
23 |
PDB header:rna binding protein
Chain: B: PDB Molecule:type iv crispr associated cas6 rna endonuclease;
PDBTitle: type iv crispr associated rna endonuclease cas6 - apo form
|
|
|
|
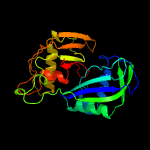
| 6 | c3i4hX_
|
|
 |
99.9 |
20 |
PDB header:hydrolase
Chain: X: PDB Molecule:endoribonuclease;
PDBTitle: crystal structure of cas6 in pyrococcus furiosus
|
|
|
|
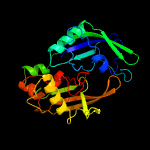
| 7 | c3qjjB_
|
|
 |
99.9 |
14 |
PDB header:immune system/rna
Chain: B: PDB Molecule:putative uncharacterized protein ph0350;
PDBTitle: one ramp protein binding different rna substrates
|
|
|
|
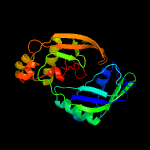
| 8 | c5yi6A_
|
|
 |
99.9 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:crispr-associated endoribonuclease cas6 1;
PDBTitle: crispr associated protein cas6
|
|
|
|
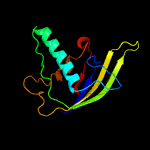
| 9 | c6dd5B_
|
|
 |
98.1 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:mmb-1 cas6 fused to maltose binding protein,crispr-
PDBTitle: crystal structure of the cas6 domain of marinomonas mediterranea mmb-12 cas6-rt-cas1 fusion protein
|
|
|
|
| 10 | c4z7lB_
|
|
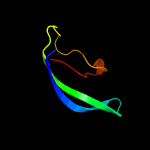 |
96.2 |
19 |
PDB header:hydrolase/rna
Chain: B: PDB Molecule:cas6b;
PDBTitle: crystal structure of cas6b
|
|
|
|
| 11 | c6ae2B_
|
|
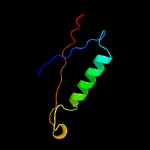 |
95.3 |
17 |
PDB header:rna binding protein
Chain: B: PDB Molecule:csm3;
PDBTitle: crystal structure of csm3 of the type iii-a crispr-cas effector2 complex
|
|
|
|
| 12 | c4l6uB_
|
|
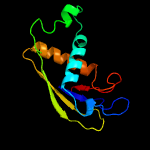 |
94.8 |
21 |
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of af1868: cmr1 subunit of the cmr rna silencing2 complex
|
|
|
|
| 13 | c4n0lB_
|
|
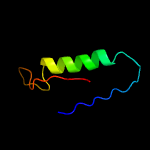 |
94.7 |
21 |
PDB header:rna binding protein
Chain: B: PDB Molecule:predicted component of a thermophile-specific dna repair
PDBTitle: methanopyrus kandleri csm3 crystal structure
|
|
|
|
| 14 | c5yjdB_
|
|
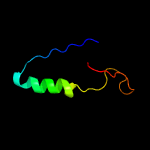 |
94.7 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:csm3;
PDBTitle: structural insights into the crispr-cas-associated ribonuclease2 activity of staphylococcus epidermidis csm3
|
|
|
|
| 15 | c4qtsC_
|
|
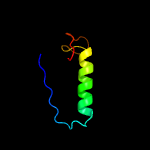 |
94.0 |
18 |
PDB header:rna binding protein
Chain: C: PDB Molecule:crispr type iii-associated ramp protein csm3;
PDBTitle: crystal structure of csm3-csm4 subcomplex in the type iii-a crispr-cas2 interference complex
|
|
|
|
| 16 | c4w8vA_
|
|
 |
93.3 |
15 |
PDB header:rna binding protein
Chain: A: PDB Molecule:crispr system cmr subunit cmr6;
PDBTitle: crystal structure of cmr6 from pyrococcus furiosus
|
|
|
|
| 17 | c4w8wB_
|
|
 |
93.3 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:crispr system cmr subunit cmr4;
PDBTitle: crystal structure of oligomeric cmr4 from pyrococcus furiosus
|
|
|
|
| 18 | c6muuE_
|
|
 |
93.1 |
23 |
PDB header:rna binding protein/rna
Chain: E: PDB Molecule:uncharacterized protein csm4;
PDBTitle: cryo-em structure of csm-crrna binary complex in type iii-a crispr-cas2 system
|
|
|
|
| 19 | c6ifnF_
|
|
 |
92.9 |
18 |
PDB header:rna binding protein
Chain: F: PDB Molecule:type iii-a crispr-associated ramp protein csm3;
PDBTitle: crystal structure of type iii-a crispr csm complex
|
|
|
|
| 20 | c6musK_
|
|
 |
92.8 |
23 |
PDB header:rna binding protein/rna
Chain: K: PDB Molecule:uncharacterized protein csm3;
PDBTitle: cryo-em structure of larger csm-crrna-target rna ternary complex in2 type iii-a crispr-cas system
|
|
|
|
| 21 | c4w8xA_ |
|
not modelled |
92.0 |
12 |
PDB header:rna binding protein
Chain: A: PDB Molecule:crispr system cmr subunit cmr1-1;
PDBTitle: crystal structure of cmr1 from pyrococcus furiosus bound to a2 nucleotide
|
|
|
| 22 | c3x1lH_ |
|
not modelled |
87.6 |
24 |
PDB header:rna binding protein/rna/dna
Chain: H: PDB Molecule:cmr6;
PDBTitle: crystal structure of the crispr-cas rna silencing cmr complex bound to2 a target analog
|
|
|
| 23 | c4qtsB_ |
|
not modelled |
82.1 |
10 |
PDB header:rna binding protein
Chain: B: PDB Molecule:crispr type iii-associated ramp protein csm4;
PDBTitle: crystal structure of csm3-csm4 subcomplex in the type iii-a crispr-cas2 interference complex
|
|
|
| 24 | c3x1lE_ |
|
not modelled |
77.3 |
20 |
PDB header:rna binding protein/rna/dna
Chain: E: PDB Molecule:cmr4;
PDBTitle: crystal structure of the crispr-cas rna silencing cmr complex bound to2 a target analog
|
|
|
| 25 | c6ig0B_ |
|
not modelled |
65.8 |
24 |
PDB header:rna binding protein
Chain: B: PDB Molecule:type iii-a crispr-associated ramp protein csm4;
PDBTitle: type iii-a csm complex, cryo-em structure of csm-ctr1, atp bound
|
|
|
| 26 | c3qrqA_ |
|
not modelled |
62.2 |
21 |
PDB header:rna binding protein/rna
Chain: A: PDB Molecule:putative uncharacterized protein tthb192;
PDBTitle: structure of thermus thermophilus cse3 bound to an rna representing a2 pre-cleavage complex
|
|
|
| 27 | c4dzdA_ |
|
not modelled |
61.3 |
35 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein ygch;
PDBTitle: crystal structure of the crispr-associated protein cas6e from2 escherichia coli str. k-12
|
|
|
| 28 | c1wj9A_ |
|
not modelled |
48.5 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:crispr-associated protein;
PDBTitle: crystal structure of a crispr-associated protein from2 thermus thermophilus
|
|
|
| 29 | d1wj9a2 |
|
not modelled |
47.8 |
24 |
Fold:Ferredoxin-like
Superfamily:CRISPR-associated protein
Family:CRISPR-associated protein |
|
|
| 30 | c5u0aA_ |
|
not modelled |
32.1 |
20 |
PDB header:immune system
Chain: A: PDB Molecule:crispr-associated protein, cse3 family;
PDBTitle: crispr rna-guided surveillance complex
|
|
|
| 31 | d1cuka2 |
|
not modelled |
30.1 |
16 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:DNA helicase RuvA subunit, middle domain |
|
|
| 32 | d1ixra1 |
|
not modelled |
26.8 |
24 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:DNA helicase RuvA subunit, middle domain |
|
|
| 33 | d1bvsa2 |
|
not modelled |
26.3 |
28 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:DNA helicase RuvA subunit, middle domain |
|
|
| 34 | c5xe3F_ |
|
not modelled |
22.5 |
37 |
PDB header:hydrolase/antitoxin
Chain: F: PDB Molecule:probable antitoxin maze4;
PDBTitle: endoribonuclease in complex with its cognate antitoxin from2 mycobacterial species
|
|
|
| 35 | c5xe3E_ |
|
not modelled |
22.5 |
37 |
PDB header:hydrolase/antitoxin
Chain: E: PDB Molecule:probable antitoxin maze4;
PDBTitle: endoribonuclease in complex with its cognate antitoxin from2 mycobacterial species
|
|
|
| 36 | d1pzqa_ |
|
not modelled |
21.2 |
22 |
Fold:Dimerisation interlock
Superfamily:Docking domain A of the erythromycin polyketide synthase (DEBS)
Family:Docking domain A of the erythromycin polyketide synthase (DEBS) |
|
|
| 37 | c1xmeB_ |
|
not modelled |
20.2 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome c oxidase polypeptide ii;
PDBTitle: structure of recombinant cytochrome ba3 oxidase from thermus2 thermophilus
|
|
|
| 38 | c1ixrB_ |
|
not modelled |
19.5 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:holliday junction dna helicase ruva;
PDBTitle: ruva-ruvb complex
|
|
|
| 39 | c2h5xA_ |
|
not modelled |
19.5 |
22 |
PDB header:dna binding protein
Chain: A: PDB Molecule:holliday junction atp-dependent dna helicase ruva;
PDBTitle: ruva from mycobacterium tuberculosis
|
|
|
| 40 | c1d8lA_ |
|
not modelled |
18.7 |
15 |
PDB header:gene regulation
Chain: A: PDB Molecule:protein (holliday junction dna helicase ruva);
PDBTitle: e. coli holliday junction binding protein ruva nh2 region lacking2 domain iii
|
|
|
| 41 | c5gmkJ_ |
|
not modelled |
17.2 |
36 |
PDB header:rna binding protein/rna
Chain: J: PDB Molecule:pre-mrna-splicing factor cwc21;
PDBTitle: cryo-em structure of the catalytic step i spliceosome (c complex) at2 3.4 angstrom resolution
|
|
|
| 42 | c6iczU_ |
|
not modelled |
14.2 |
31 |
PDB header:splicing
Chain: U: PDB Molecule:serine/arginine repetitive matrix protein 2;
PDBTitle: cryo-em structure of a human post-catalytic spliceosome (p complex) at2 3.0 angstrom
|
|
|
| 43 | c1ixrA_ |
|
not modelled |
13.9 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:holliday junction dna helicase ruva;
PDBTitle: ruva-ruvb complex
|
|
|
| 44 | c6bk8K_ |
|
not modelled |
13.2 |
31 |
PDB header:rna binding protein
Chain: K: PDB Molecule:pre-mrna-splicing factor cwc21;
PDBTitle: s. cerevisiae spliceosomal post-catalytic p complex
|
|
|
| 45 | c1y6uA_ |
|
not modelled |
12.9 |
8 |
PDB header:dna binding protein
Chain: A: PDB Molecule:excisionase from transposon tn916;
PDBTitle: the structure of the excisionase (xis) protein from2 conjugative transposon tn916 provides insights into the3 regulation of heterobivalent tyrosine recombinases
|
|
|
| 46 | c3btpA_ |
|
not modelled |
12.7 |
30 |
PDB header:dna binding protein, chaperone
Chain: A: PDB Molecule:single-strand dna-binding protein;
PDBTitle: crystal structure of agrobacterium tumefaciens vire2 in complex with2 its chaperone vire1: a novel fold and implications for dna binding
|
|
|
| 47 | c2wg7B_ |
|
not modelled |
12.6 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative phospholipase a2;
PDBTitle: structure of oryza sativa (rice) pla2
|
|
|
| 48 | c1hjpA_ |
|
not modelled |
12.1 |
15 |
PDB header:dna recombination
Chain: A: PDB Molecule:ruva;
PDBTitle: holliday junction binding protein ruva from e. coli
|
|
|
| 49 | d2bcqa2 |
|
not modelled |
11.6 |
28 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:DNA polymerase beta-like, second domain |
|
|
| 50 | d2vana1 |
|
not modelled |
11.6 |
17 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:DNA polymerase beta-like, second domain |
|
|
| 51 | c3tadB_ |
|
not modelled |
10.9 |
23 |
PDB header:protein binding
Chain: B: PDB Molecule:liprin-alpha-2;
PDBTitle: crystal structure of the liprin-alpha/liprin-beta complex
|
|
|
| 52 | c5mqfS_ |
|
not modelled |
10.7 |
31 |
PDB header:splicing
Chain: S: PDB Molecule:serine/arginine repetitive matrix protein 2;
PDBTitle: cryo-em structure of a human spliceosome activated for step 2 of2 splicing (c* complex)
|
|
|
| 53 | d1ux5a_ |
|
not modelled |
10.3 |
19 |
Fold:Formin homology 2 domain (FH2 domain)
Superfamily:Formin homology 2 domain (FH2 domain)
Family:Formin homology 2 domain (FH2 domain) |
|
|
| 54 | c1y64B_ |
|
not modelled |
10.3 |
19 |
PDB header:structural protein
Chain: B: PDB Molecule:bni1 protein;
PDBTitle: bni1p formin homology 2 domain complexed with atp-actin
|
|
|
| 55 | c5nocA_ |
|
not modelled |
10.1 |
9 |
PDB header:dna binding protein
Chain: A: PDB Molecule:stage 0 sporulation protein j;
PDBTitle: solution nmr structure of the c-terminal domain of parb (spo0j)
|
|
|
| 56 | d2o3aa1 |
|
not modelled |
9.8 |
45 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:AF0751-like |
|
|
| 57 | d1jmsa3 |
|
not modelled |
9.6 |
22 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:DNA polymerase beta-like, second domain |
|
|
| 58 | c5gqtA_ |
|
not modelled |
9.0 |
24 |
PDB header:plant protein
Chain: A: PDB Molecule:nitrile-specifier protein 1;
PDBTitle: crystal structure of a specifier protein from arabidopsis thaliana
|
|
|
| 59 | c6nusA_ |
|
not modelled |
8.2 |
31 |
PDB header:viral protein
Chain: A: PDB Molecule:nsp12;
PDBTitle: sars-coronavirus nsp12 bound to nsp8 co-factor
|
|
|
| 60 | d1bxua_ |
|
not modelled |
8.1 |
32 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
|
|
| 61 | d2p19a1 |
|
not modelled |
7.8 |
9 |
Fold:Chorismate lyase-like
Superfamily:Chorismate lyase-like
Family:UTRA domain |
|
|
| 62 | c4tvxU_ |
|
not modelled |
7.7 |
16 |
|
| 63 | c5mqcA_ |
|
not modelled |
7.5 |
63 |
PDB header:virus
Chain: A: PDB Molecule:vp1;
PDBTitle: structure of black queen cell virus
|
|
|
| 64 | d1or7c_ |
|
not modelled |
7.5 |
12 |
Fold:N-terminal, cytoplasmic domain of anti-sigmaE factor RseA
Superfamily:N-terminal, cytoplasmic domain of anti-sigmaE factor RseA
Family:N-terminal, cytoplasmic domain of anti-sigmaE factor RseA |
|
|
| 65 | c1or7C_ |
|
not modelled |
7.5 |
12 |
PDB header:transcription
Chain: C: PDB Molecule:sigma-e factor negative regulatory protein;
PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
|
|
|
| 66 | c2h1xB_ |
|
not modelled |
7.4 |
30 |
PDB header:hydrolase
Chain: B: PDB Molecule:5-hydroxyisourate hydrolase (formerly known as
PDBTitle: crystal structure of 5-hydroxyisourate hydrolase (formerly2 known as trp, transthyretin related protein)
|
|
|
| 67 | c3ggeA_ |
|
not modelled |
7.4 |
7 |
PDB header:protein binding
Chain: A: PDB Molecule:pdz domain-containing protein gipc2;
PDBTitle: crystal structure of the pdz domain of pdz domain-containing protein2 gipc2
|
|
|
| 68 | d1oo2a_ |
|
not modelled |
7.3 |
32 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
|
|
| 69 | d1b26a2 |
|
not modelled |
7.1 |
14 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
|
|
| 70 | c6dxwC_ |
|
not modelled |
7.0 |
8 |
PDB header:hydrolase
Chain: C: PDB Molecule:n-acylethanolamine-hydrolyzing acid amidase;
PDBTitle: human n-acylethanolamine-hydrolyzing acid amidase (naaa) precursor2 (c126a)
|
|
|
| 71 | c4jhdF_ |
|
not modelled |
6.8 |
67 |
PDB header:structural protein/protein binding
Chain: F: PDB Molecule:protein cordon-bleu;
PDBTitle: crystal structure of an actin dimer in complex with the actin2 nucleator cordon-bleu
|
|
|
| 72 | c4wwxB_ |
|
not modelled |
6.6 |
13 |
PDB header:hydrolase, ligase
Chain: B: PDB Molecule:v(d)j recombination-activating protein 1;
PDBTitle: crystal structure of the core rag1/2 recombinase
|
|
|
| 73 | c1pggB_ |
|
not modelled |
6.5 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prostaglandin h2 synthase-1;
PDBTitle: prostaglandin h2 synthase-1 complexed with 1-(4-iodobenzoyl)-5-2 methoxy-2-methylindole-3-acetic acid (iodoindomethacin), trans model
|
|
|
| 74 | c1ht8B_ |
|
not modelled |
6.5 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prostaglandin h2 synthase-1;
PDBTitle: the 2.7 angstrom resolution model of ovine cox-1 complexed with2 alclofenac
|
|
|
| 75 | d1tfpa_ |
|
not modelled |
6.4 |
36 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
|
|
| 76 | c6ir9W_ |
|
not modelled |
6.3 |
10 |
PDB header:transcription/rna/dna
Chain: W: PDB Molecule:spt5;
PDBTitle: rna polymerase ii elongation complex bound with elf1 and spt4/5,2 stalled at shl(-1) of the nucleosome
|
|
|
| 77 | d1f86a_ |
|
not modelled |
6.2 |
30 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
|
|
| 78 | d1kgia_ |
|
not modelled |
6.2 |
32 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
|
|
| 79 | c5cdcA_ |
|
not modelled |
6.2 |
50 |
PDB header:virus
Chain: A: PDB Molecule:vp1, structural polyprotein;
PDBTitle: crystal structure of israel acute paralysis virus
|
|
|
| 80 | c6h3jA_ |
|
not modelled |
6.0 |
22 |
PDB header:protein transport
Chain: A: PDB Molecule:protein involved in gliding motility spra;
PDBTitle: structural snapshots of the type 9 protein translocon plug-complex
|
|
|
| 81 | c2n5qA_ |
|
not modelled |
6.0 |
27 |
PDB header:unknown function
Chain: A: PDB Molecule:cysteine-rich peptide js1;
PDBTitle: solution structure of cystein-rich peptide js1 from jasminum sambac
|
|
|
| 82 | d1cmwa1 |
|
not modelled |
6.0 |
30 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
|
|
| 83 | c2oyuP_ |
|
not modelled |
5.8 |
14 |
PDB header:oxidoreductase
Chain: P: PDB Molecule:prostaglandin g/h synthase 1;
PDBTitle: indomethacin-(s)-alpha-ethyl-ethanolamide bound to cyclooxygenase-1
|
|
|
| 84 | c1ddxA_ |
|
not modelled |
5.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (prostaglandin h2 synthase-2);
PDBTitle: crystal structure of a mixture of arachidonic acid and prostaglandin2 bound to the cyclooxygenase active site of cox-2: prostaglandin3 structure
|
|
|
| 85 | d1ttaa_ |
|
not modelled |
5.6 |
30 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
|
|
| 86 | c3a0hJ_ |
|
not modelled |
5.6 |
31 |
PDB header:electron transport
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: crystal structure of i-substituted photosystem ii complex
|
|
|
| 87 | d2axtj1 |
|
not modelled |
5.6 |
31 |
Fold:Single transmembrane helix
Superfamily:Photosystem II reaction center protein J, PsbJ
Family:PsbJ-like |
|
|
| 88 | d1na6a1 |
|
not modelled |
5.5 |
27 |
Fold:DNA-binding pseudobarrel domain
Superfamily:DNA-binding pseudobarrel domain
Family:Type II restriction endonuclease effector domain |
|
|
| 89 | c3ui3A_ |
|
not modelled |
5.5 |
20 |
PDB header:rna binding protein
Chain: A: PDB Molecule:immunoglobulin g-binding protein g, virulence-associated
PDBTitle: structural and biochemical characterization of hp0315 from2 helicobacter pylori as a vapd protein with an endoribonuclease3 activity
|
|
|
| 90 | d1mc8a1 |
|
not modelled |
5.5 |
38 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
|
|
| 91 | c4luqB_ |
|
not modelled |
5.5 |
19 |
PDB header:protein binding/toxin inhibitor
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of virulence effector tse3 in complex with2 neutralizer tsi3
|
|
|
| 92 | d2fmpa2 |
|
not modelled |
5.4 |
22 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:DNA polymerase beta-like, second domain |
|
|
| 93 | c4khbE_ |
|
not modelled |
5.4 |
30 |
PDB header:transcription/replication
Chain: E: PDB Molecule:uncharacterized protein spt16d;
PDBTitle: structure of the spt16d pob3n heterodimer
|
|
|
| 94 | c5ot4D_ |
|
not modelled |
5.4 |
47 |
PDB header:transport protein
Chain: D: PDB Molecule:interaptin;
PDBTitle: structure of the legionella pneumophila effector ridl (1-866)
|
|
|
| 95 | c6iicA_ |
|
not modelled |
5.3 |
25 |
PDB header:virus
Chain: A: PDB Molecule:vp1 of mud crab dicistrovirus;
PDBTitle: cryoem structure of mud crab dicistrovirus
|
|
|
| 96 | c2gpzC_ |
|
not modelled |
5.3 |
29 |
PDB header:hydrolase
Chain: C: PDB Molecule:transthyretin-like protein;
PDBTitle: transthyretin-like protein from salmonella dublin
|
|
|
| 97 | c3qvaB_ |
|
not modelled |
5.2 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:transthyretin-like protein;
PDBTitle: structure of klebsiella pneumoniae 5-hydroxyisourate hydrolase
|
|
|
| 98 | c6h3iA_ |
|
not modelled |
5.1 |
22 |
PDB header:protein transport
Chain: A: PDB Molecule:protein involved in gliding motility spra;
PDBTitle: structural snapshots of the type 9 protein translocon
|
|
|
| 99 | c4atoA_ |
|
not modelled |
5.1 |
29 |
PDB header:toxin/antitoxin
Chain: A: PDB Molecule:toxn;
PDBTitle: new insights into the mechanism of bacterial type iii toxin-antitoxin2 systems: selective toxin inhibition by a non-coding rna pseudoknot
|
|
|