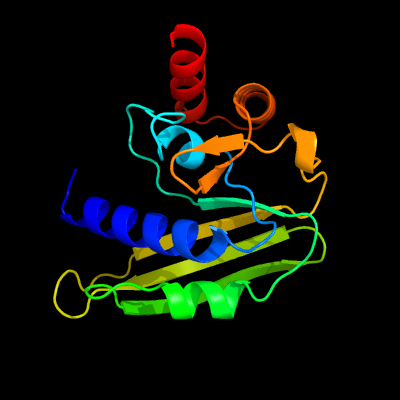
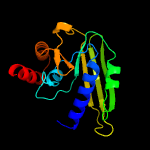
1 c4ok0B_
99.5
21
PDB header: transferaseChain: B: PDB Molecule: putative;PDBTitle: crystal structure of putative nucleotidyltransferase from h. pylori
2 d1h9aa2
54.2
48
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Glucose 6-phosphate dehydrogenase-like
3 d1qkia2
42.8
43
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Glucose 6-phosphate dehydrogenase-like
4 c1h9aA_
42.7
48
PDB header: oxidoreductase (choh(d) - nad(p))Chain: A: PDB Molecule: glucose 6-phosphate 1-dehydrogenase;PDBTitle: complex of active mutant (q365->c) of glucose 6-phosphate2 dehydrogenase from l. mesenteroides with coenzyme nadp
5 c4e9iB_
40.8
43
PDB header: oxidoreductaseChain: B: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: glucose-6-p dehydrogenase (apo form) from trypanosoma cruzi
6 c5xw4A_
39.6
24
PDB header: cell cycleChain: A: PDB Molecule: tyrosine-protein phosphatase cdc14;PDBTitle: crystal structure of budding yeast cdc14p (wild type) in the apo state
7 c4lgvA_
38.5
52
PDB header: oxidoreductaseChain: A: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: x-ray crystal structure of glucose-6-phosphate 1-dehydrogenase from2 mycobacterium avium
8 c2bhlB_
37.7
43
PDB header: oxidoreductaseChain: B: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: x-ray structure of human glucose-6-phosphate dehydrogenase (deletion2 variant) complexed with glucose-6-phosphate
9 c1qkiE_
37.0
43
PDB header: oxidoreductaseChain: E: PDB Molecule: glucose-6-phosphate 1-dehydrogenase;PDBTitle: x-ray structure of human glucose 6-phosphate dehydrogenase (variant2 canton r459l) complexed with structural nadp+
10 c4dzdA_
36.7
45
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein ygch;PDBTitle: crystal structure of the crispr-associated protein cas6e from2 escherichia coli str. k-12
11 d1wj9a2
35.3
45
Fold: Ferredoxin-likeSuperfamily: CRISPR-associated proteinFamily: CRISPR-associated protein
12 c3qrqA_
33.7
45
PDB header: rna binding protein/rnaChain: A: PDB Molecule: putative uncharacterized protein tthb192;PDBTitle: structure of thermus thermophilus cse3 bound to an rna representing a2 pre-cleavage complex
13 c1wj9A_
32.6
45
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: crispr-associated protein;PDBTitle: crystal structure of a crispr-associated protein from2 thermus thermophilus
14 c5u0aA_
30.3
43
PDB header: immune systemChain: A: PDB Molecule: crispr-associated protein, cse3 family;PDBTitle: crispr rna-guided surveillance complex
15 d1fpza_
27.1
24
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
16 c1fpzF_
21.5
24
PDB header: hydrolaseChain: F: PDB Molecule: cyclin-dependent kinase inhibitor 3;PDBTitle: crystal structure analysis of kinase associated phosphatase (kap) with2 a substitution of the catalytic site cysteine (cys140) to a serine
17 c4nyhB_
18.7
30
PDB header: hydrolaseChain: B: PDB Molecule: rna/rnp complex-1-interacting phosphatase;PDBTitle: orthorhombic crystal form of pir1 dual specificity phosphatase core
18 c1yn9B_
15.8
24
PDB header: hydrolaseChain: B: PDB Molecule: polynucleotide 5'-phosphatase;PDBTitle: crystal structure of baculovirus rna 5'-phosphatase2 complexed with phosphate
19 c5z5bA_
12.3
32
PDB header: hydrolaseChain: A: PDB Molecule: protein-tyrosine phosphatase;PDBTitle: crystal structure of tk-ptp in the g95a mutant form
20 d1j2oa2
10.8
73
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain
21 c2c46B_
not modelled
10.1
23
PDB header: transferaseChain: B: PDB Molecule: mrna capping enzyme;PDBTitle: crystal structure of the human rna guanylyltransferase and 5'-2 phosphatase
22 c4ps2A_
not modelled
9.5
36
PDB header: contractile proteinChain: A: PDB Molecule: putative type vi secretion protein;PDBTitle: structure of the c-terminal fragment (87-165) of e.coli eaec tssb2 molecule
23 d1dgsa3
not modelled
9.1
45
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: Adenylation domain of NAD+-dependent DNA ligase
24 c2ltuA_
not modelled
9.1
53
PDB header: transferaseChain: A: PDB Molecule: 5'-amp-activated protein kinase catalytic subunit alpha-2;PDBTitle: solution structure of autoinhibitory domain of human amp-activated2 protein kinase catalytic subunit
25 c6bmnA_
not modelled
8.9
36
PDB header: transferaseChain: A: PDB Molecule: human dhhc20 palmitoyltransferase;PDBTitle: structure of human dhhc20 palmitoyltransferase, space group p63
26 c1dvpA_
not modelled
8.8
26
PDB header: transferaseChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosinePDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
27 c5lo2A_
not modelled
8.7
45
PDB header: structural proteinChain: A: PDB Molecule: ppatyr;PDBTitle: engineering protein stability with atomic precision in a monomeric2 miniprotein
28 c3ld0Q_
not modelled
8.5
50
PDB header: gene regulationChain: Q: PDB Molecule: inhibitor of trap, regulated by t-box (trp) sequence rtpa;PDBTitle: crystal structure of b.licheniformis anti-trap protein, an antagonist2 of trap-rna interactions
29 c1yx5A_
not modelled
8.0
46
PDB header: hydrolaseChain: A: PDB Molecule: 26s proteasome non-atpase regulatory subunit 4;PDBTitle: solution structure of s5a uim-1/ubiquitin complex
30 d1r7aa1
not modelled
7.9
70
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
31 c1dgsB_
not modelled
7.7
45
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase from t.2 filiformis
32 c2ly8A_
not modelled
7.2
24
PDB header: chaperoneChain: A: PDB Molecule: budding yeast chaperone scm3;PDBTitle: the budding yeast chaperone scm3 recognizes the partially unfolded2 dimer of the centromere-specific cse4/h4 histone variant
33 c5o60P_
not modelled
6.8
40
PDB header: ribosomeChain: P: PDB Molecule: 50s ribosomal protein l18;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
34 c3j3vO_
not modelled
6.6
47
PDB header: ribosomeChain: O: PDB Molecule: 50s ribosomal protein l18;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
35 d2oa5a1
not modelled
6.4
35
Fold: BLRF2-likeSuperfamily: BLRF2-likeFamily: BLRF2-like
36 c3bboQ_
not modelled
6.3
47
PDB header: ribosomeChain: Q: PDB Molecule: ribosomal protein l18;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
37 c5lo4A_
not modelled
6.2
40
PDB header: structural proteinChain: A: PDB Molecule: ppa-ch3;PDBTitle: engineering protein stability with atomic precision in a monomeric2 miniprotein
38 c6eonA_
not modelled
5.8
25
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: galactanase bt0290
39 d1d5ra2
not modelled
5.7
24
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like
40 d1cixa_
not modelled
5.7
71
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins
41 c1cixA_
not modelled
5.7
71
PDB header: antimicrobial peptideChain: A: PDB Molecule: protein (tachystatin a);PDBTitle: three-dimensional structure of antimicrobial peptide2 tachystatin a isolated from horseshoe crab
42 d1elka_
not modelled
5.6
26
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain
43 d2r7da1
not modelled
5.5
73
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
44 c6mv5P_
not modelled
5.3
44
PDB header: immune systemChain: P: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: anti-pcsk9 fab 6e2 bound to the n-terminal peptide from pcsk9 (e32k)
45 c5j9hA_
not modelled
5.2
29
PDB header: viral proteinChain: A: PDB Molecule: envelopment polyprotein;PDBTitle: crystal structure of glycoprotein c from puumala virus in the post-2 fusion conformation (ph 8.0)
46 c4a1sE_
not modelled
5.2
100
PDB header: cell cycleChain: E: PDB Molecule: re60102p;PDBTitle: crystallographic structure of the pins:insc complex
47 c1sseA_
not modelled
5.1
43
PDB header: transcription activatorChain: A: PDB Molecule: ap-1 like transcription factor yap1;PDBTitle: solution structure of the oxidized form of the yap1 redox2 domain
48 c2bc4C_
not modelled
5.1
31
PDB header: immune systemChain: C: PDB Molecule: hla class ii histocompatibility antigen, dm alpha chain;PDBTitle: crystal structure of hla-dm