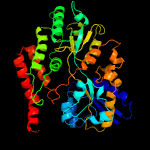
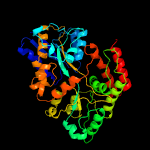
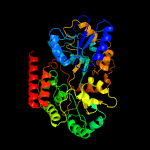
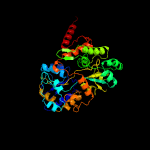
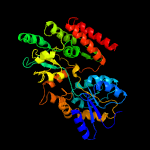
1 c4mfiA_
100.0
100
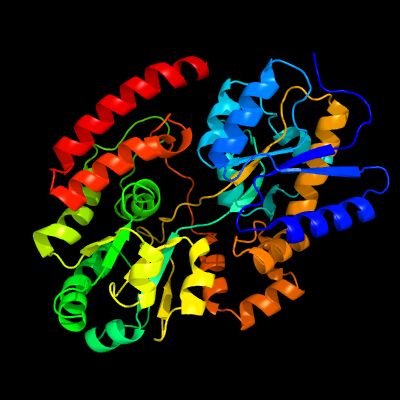
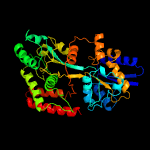
PDB header: sugar binding proteinChain: A: PDB Molecule: sn-glycerol-3-phosphate abc transporter substrate-bindingPDBTitle: crystal structure of mycobacterium tuberculosis ugpb
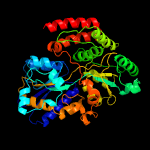
2 c4aq4A_
100.0
23
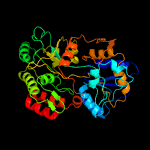
PDB header: diester-binding proteinChain: A: PDB Molecule: sn-glycerol-3-phosphate-binding periplasmic protein ugpb;PDBTitle: substrate bound sn-glycerol-3-phosphate binding periplasmic protein2 ugpb from escherichia coli
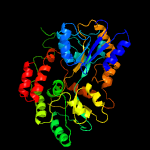
3 c6dtqC_
100.0
22
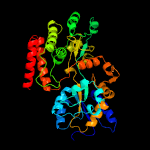
PDB header: sugar binding proteinChain: C: PDB Molecule: maltose-binding protein male3;PDBTitle: maltose bound t. maritima male3
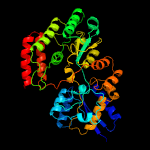
4 c3uorB_
100.0
19
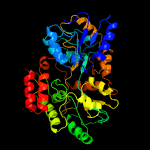
PDB header: sugar binding proteinChain: B: PDB Molecule: abc transporter sugar binding protein;PDBTitle: the structure of the sugar-binding protein male from the phytopathogen2 xanthomonas citri
5 d1eu8a_
100.0
20
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
6 c3osqA_
100.0
17
PDB header: fluorescent protein, transport proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,green fluorescentPDBTitle: maltose-bound maltose sensor engineered by insertion of circularly2 permuted green fluorescent protein into e. coli maltose binding3 protein at position 175
7 c4r6kA_
100.0
12
PDB header: transport proteinChain: A: PDB Molecule: solute-binding protein;PDBTitle: crystal structure of abc transporter substrate-binding protein yeso2 from bacillus subtilis, target efi-510761, an open conformation
8 c3k02A_
100.0
20
PDB header: transport proteinChain: A: PDB Molecule: acarbose/maltose binding protein gach;PDBTitle: crystal structures of the gach receptor of streptomyces glaucescens2 gla.o in the unliganded form and in complex with acarbose and an3 acarbose homolog. comparison with acarbose-loaded maltose binding4 protein of salmonella typhimurium.
9 c5azaA_
100.0
16
PDB header: sugar binding protein, transferaseChain: A: PDB Molecule: maltose-binding periplasmic protein,oligosaccharylPDBTitle: crystal structure of mbp-saglb fusion protein with a 20-residue spacer2 in the connector helix
10 c3h4zC_
100.0
16
PDB header: allergenChain: C: PDB Molecule: maltose-binding periplasmic protein fused with allergenPDBTitle: crystal structure of an mbp-der p 7 fusion protein
11 c6aeoA_
100.0
15
PDB header: protein transportChain: A: PDB Molecule: maltose/maltodextrin-binding periplasmic protein,tssl;PDBTitle: tssl periplasmic domain
12 c4gqoC_
100.0
16
PDB header: unknown functionChain: C: PDB Molecule: lmo0859 protein;PDBTitle: 2.1 angstrom resolution crystal structure of uncharacterized protein2 lmo0859 from listeria monocytogenes egd-e
13 c4ry1A_
100.0
17
PDB header: transport proteinChain: A: PDB Molecule: periplasmic solute binding protein;PDBTitle: crystal structure of periplasmic solute binding protein eca2210 from2 pectobacterium atrosepticum scri1043, target efi-510858
14 c4wrnB_
100.0
16
PDB header: structural proteinChain: B: PDB Molecule: maltose-binding periplasmic protein,uromodulin;PDBTitle: crystal structure of the polymerization region of human2 uromodulin/tamm-horsfall protein
15 c5i04A_
100.0
16
PDB header: signaling proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,endoglin;PDBTitle: crystal structure of the orphan region of human endoglin/cd105
16 c2uvgA_
100.0
15
PDB header: sugar-binding proteinChain: A: PDB Molecule: abc type periplasmic sugar-binding protein;PDBTitle: structure of a periplasmic oligogalacturonide binding2 protein from yersinia enterocolitica
17 c4h1gA_
100.0
17
PDB header: motor proteinChain: A: PDB Molecule: maltose binding protein-cakar3 motor domain fusion protein;PDBTitle: structure of candida albicans kar3 motor domain fused to maltose-2 binding protein
18 c5gxtA_
100.0
16
PDB header: protein transportChain: A: PDB Molecule: maltose-binding periplasmic protein,pigg;PDBTitle: crystal structure of pigg
19 c5hzvA_
100.0
16
PDB header: signaling proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,endoglin;PDBTitle: crystal structure of the zona pellucida module of human endoglin/cd105
20 c5k2xA_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: sugar abc transporter permease;PDBTitle: crystal structure of m. tuberculosis uspc (tetragonal crystal form)
21 c5t0aB_
not modelled
100.0
17
PDB header: transferaseChain: B: PDB Molecule: maltose binding protein - heparan sulfate 6-o-PDBTitle: crystal structure of heparan sulfate 6-o-sulfotransferase with bound2 pap and heptasaccharide substrate
22 c6dd5B_
not modelled
100.0
16
PDB header: hydrolaseChain: B: PDB Molecule: mmb-1 cas6 fused to maltose binding protein,crispr-PDBTitle: crystal structure of the cas6 domain of marinomonas mediterranea mmb-12 cas6-rt-cas1 fusion protein
23 c3ob4A_
not modelled
100.0
17
PDB header: allergenChain: A: PDB Molecule: maltose abc transporter periplasmic protein, arah 2;PDBTitle: mbp-fusion protein of the major peanut allergen ara h 2
24 c4xa2A_
not modelled
100.0
16
PDB header: cell adhesionChain: A: PDB Molecule: maltose-binding periplasmic protein,mbp-pila: c;PDBTitle: structure of the major type iv pilin of acinetobacter baumannii
25 c5fsgA_
not modelled
100.0
16
PDB header: viral proteinChain: A: PDB Molecule: maltose-binding periplasmic protein, hantavirusPDBTitle: structure of the hantavirus nucleoprotein provides insights2 into the mechanism of rna encapsidation and a template for3 drug design
26 c4kegA_
not modelled
100.0
17
PDB header: lipid binding proteinChain: A: PDB Molecule: maltose-binding periplasmic/palate lung and nasalPDBTitle: crystal structure of mbp fused human splunc1
27 c4ovjA_
not modelled
100.0
13
PDB header: transport proteinChain: A: PDB Molecule: extracellular solute-binding protein family 1;PDBTitle: extracellular solute-binding protein family 1 from alicyclobacillus2 acidocaldarius subsp. acidocaldarius dsm 446
28 c3f5fA_
not modelled
100.0
17
PDB header: transport, transferaseChain: A: PDB Molecule: maltose-binding periplasmic protein, heparan sulfate 2-o-PDBTitle: crystal structure of heparan sulfate 2-o-sulfotransferase from gallus2 gallus as a maltose binding protein fusion.
29 c4ryaA_
not modelled
100.0
16
PDB header: transport proteinChain: A: PDB Molecule: abc transporter substrate binding protein (sorbitol);PDBTitle: crystal structure of abc transporter solute binding protein avi_35672 from agrobacterium vitis s4, target efi-510645, with bound d-mannitol
30 c4egcA_
not modelled
100.0
17
PDB header: transcription/hydrolaseChain: A: PDB Molecule: maltose-binding periplasmic protein, homeobox protein six1PDBTitle: crystal structure of mbp-fused human six1 bound to human eya2 eya2 domain
31 c5eduB_
not modelled
100.0
16
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: maltose-binding periplasmic protein, histone deacetylase 6PDBTitle: crystal structure of human histone deacetylase 6 catalytic domain 2 in2 complex with trichostatin a
32 c5jqeA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: sugar abc transporter substrate-binding protein,caspase-8PDBTitle: crystal structure of caspase8 tded
33 c3py7A_
not modelled
100.0
16
PDB header: viral proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,paxillin ld1,protein e6PDBTitle: crystal structure of full-length bovine papillomavirus oncoprotein e62 in complex with ld1 motif of paxillin at 2.3a resolution
34 c3waiA_
not modelled
100.0
17
PDB header: transferase, transport proteinChain: A: PDB Molecule: maltose-binding periplasmic protein, transmembranePDBTitle: crystal structure of the c-terminal globular domain of2 oligosaccharyltransferase (afaglb-l, o29867_arcfu) from archaeoglobus3 fulgidus as a mbp fusion
35 c5yseB_
not modelled
100.0
17
PDB header: sugar binding proteinChain: B: PDB Molecule: lin1841 protein;PDBTitle: crystal structure of beta-1,2-glucooligosaccharide binding protein in2 complex with sophorotetraose
36 c5wvmA_
not modelled
100.0
16
PDB header: sugar binding proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,two-component systemPDBTitle: crystal structure of baes cocrystallized with 2 mm indole
37 c3dm0A_
not modelled
100.0
16
PDB header: sugar binding protein,signaling proteinChain: A: PDB Molecule: maltose-binding periplasmic protein fused with rack1;PDBTitle: maltose binding protein fusion with rack1 from a. thaliana
38 c5f7vA_
not modelled
100.0
14
PDB header: cycloalternan binding proteinChain: A: PDB Molecule: lmo0181 protein;PDBTitle: abc substrate-binding protein lmo0181 from listeria monocytogenes in2 complex with cycloalternan
39 c6anvA_
not modelled
100.0
16
PDB header: immune systemChain: A: PDB Molecule: anti-crispr protein acrf1 fused with c-terminal mbp tag;PDBTitle: crystal structure of anti-crispr protein acrf1
40 c4wgiA_
not modelled
100.0
17
PDB header: apoptosis/inhibitorChain: A: PDB Molecule: maltose-binding periplasmic protein,induced myeloidPDBTitle: a single diastereomer of a macrolactam core binds specifically to2 myeloid cell leukemia 1 (mcl1)
41 c4r9fA_
not modelled
100.0
17
PDB header: sugar binding proteinChain: A: PDB Molecule: mbp1;PDBTitle: cpmnbp1 with mannobiose bound
42 c3c4mA_
not modelled
100.0
16
PDB header: membrane proteinChain: A: PDB Molecule: fusion protein of maltose-binding periplasmic protein andPDBTitle: structure of human parathyroid hormone in complex with the2 extracellular domain of its g-protein-coupled receptor (pth1r)
43 c4qvhA_
not modelled
100.0
17
PDB header: transferaseChain: A: PDB Molecule: maltose-binding periplasmic protein, 4'-phosphopantetheinylPDBTitle: crystal structure of the essential mycobacterium tuberculosis2 phosphopantetheinyl transferase pptt, solved as a fusion protein with3 maltose binding protein
44 c3d4cA_
not modelled
100.0
17
PDB header: cell adhesionChain: A: PDB Molecule: maltose-binding periplasmic protein, linker, zona pellucidaPDBTitle: zp-n domain of mammalian sperm receptor zp3 (crystal form i)
45 c5tibA_
not modelled
100.0
16
PDB header: lipid binding proteinChain: A: PDB Molecule: sugar abc transporter substrate-binding protein,gasdermin-PDBTitle: gasdermin-b c-terminal domain containing the polymorphism residues2 arg299:ser306 fused to maltose binding protein
46 c5ci5B_
not modelled
100.0
18
PDB header: sugar binding proteinChain: B: PDB Molecule: extracellular solute-binding protein family 1;PDBTitle: crystal structure of an abc transporter solute binding protein from2 thermotoga lettingae tmo (tlet_1705, target efi-510544) bound with3 alpha-d-tagatose
47 c4r6hA_
not modelled
100.0
13
PDB header: transport proteinChain: A: PDB Molecule: solute binding protein msme;PDBTitle: crystal structure of putative binding protein msme from bacillus2 subtilis subsp. subtilis str. 168, target efi-510764, an open3 conformation
48 c5dfmB_
not modelled
100.0
16
PDB header: nuclear proteinChain: B: PDB Molecule: maltose-binding periplasmic protein,telomerase-associatedPDBTitle: structure of tetrahymena telomerase p19 fused to mbp
49 c4g68A_
not modelled
100.0
13
PDB header: transport proteinChain: A: PDB Molecule: abc transporter;PDBTitle: biochemical and structural insights into xylan utilization by the2 thermophilic bacteriumcaldanaerobius polysaccharolyticus
50 c1hsjA_
not modelled
100.0
16
PDB header: transcription/sugar binding proteinChain: A: PDB Molecule: fusion protein consisting of staphylococcus accessoryPDBTitle: sarr mbp fusion structure
51 c4g68C_
not modelled
100.0
13
PDB header: transport proteinChain: C: PDB Molecule: abc transporter;PDBTitle: biochemical and structural insights into xylan utilization by the2 thermophilic bacteriumcaldanaerobius polysaccharolyticus
52 c5ii5A_
not modelled
100.0
16
PDB header: cell adhesionChain: A: PDB Molecule: maltose-binding periplasmic protein,vitelline envelopePDBTitle: crystal structure of red abalone verl repeat 1 at 1.8 a resolution
53 c2z8fB_
not modelled
100.0
15
PDB header: sugar binding proteinChain: B: PDB Molecule: galacto-n-biose/lacto-n-biose i transporter substrate-PDBTitle: the galacto-n-biose-/lacto-n-biose i-binding protein (gl-bp) of the2 abc transporter from bifidobacterium longum in complex with lacto-n-3 tetraose
54 c1r6zA_
not modelled
100.0
17
PDB header: gene regulationChain: A: PDB Molecule: chimera of maltose-binding periplasmic protein andPDBTitle: the crystal structure of the argonaute2 paz domain (as a mbp fusion)
55 c4r2fA_
not modelled
100.0
15
PDB header: transport proteinChain: A: PDB Molecule: extracellular solute-binding protein family 1;PDBTitle: crystal structure of sugar transporter achl_0255 from arthrobacter2 chlorophenolicus a6, target efi-510633, with bound laminaribiose
56 c5k94B_
not modelled
100.0
18
PDB header: protein transportChain: B: PDB Molecule: maltose-binding periplasmic protein,protein translocasePDBTitle: deletion-insertion chimera of mbp with the preprotein cross-linking2 domain of the seca atpase
57 c3vd8A_
not modelled
100.0
17
PDB header: sugar binding protein,signaling proteinChain: A: PDB Molecule: maltose-binding periplasmic protein, interferon-induciblePDBTitle: crystal structure of human aim2 pyd domain with mbp fusion
58 c5iaiA_
not modelled
100.0
18
PDB header: solute-binding proteinChain: A: PDB Molecule: sugar abc transporter;PDBTitle: crystal structure of abc transporter solute binding protein arad_98872 from agrobacterium radiobacter k84, target efi-510945 in complex with3 ribitol
59 c4b3nA_
not modelled
100.0
16
PDB header: sugar binding protein/ligaseChain: A: PDB Molecule: maltose-binding periplasmic protein, tripartite motif-PDBTitle: crystal structure of rhesus trim5alpha pry/spry domain
60 c3o3uN_
not modelled
100.0
17
PDB header: transport protein, signaling proteinChain: N: PDB Molecule: maltose-binding periplasmic protein, advanced glycosylationPDBTitle: crystal structure of human receptor for advanced glycation endproducts2 (rage)
61 c4tsmC_
not modelled
100.0
17
PDB header: cell adhesionChain: C: PDB Molecule: maltose-binding protein, pilin chimera;PDBTitle: mbp-fusion protein of pila1 from c. difficile r20291 residues 26-166
62 c4kv3A_
not modelled
100.0
17
PDB header: protein transportChain: A: PDB Molecule: chimera fusion protein of esx-1 secretion system proteinPDBTitle: ubiquitin-like domain of the mycobacterium tuberculosis type vii2 secretion system protein eccd1 as maltose-binding protein fusion
63 c3qufB_
not modelled
100.0
16
PDB header: transport proteinChain: B: PDB Molecule: extracellular solute-binding protein, family 1;PDBTitle: the structure of a family 1 extracellular solute-binding protein from2 bifidobacterium longum subsp. infantis
64 c4edqA_
not modelled
100.0
17
PDB header: transport protein/contractile proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,myosin-binding proteinPDBTitle: mbp-fusion protein of myosin-binding protein c residues 149-269
65 c4qszB_
not modelled
100.0
15
PDB header: transcriptionChain: B: PDB Molecule: maltose-binding periplasmic protein, jmjc domain-containingPDBTitle: crystal structure of mouse jmjd7 fused with maltose-binding protein
66 c5jonA_
not modelled
100.0
16
PDB header: transport proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,potassium/sodiumPDBTitle: crystal structure of the unliganded form of hcn2 cnbd
67 c6fflA_
not modelled
100.0
17
PDB header: sugar binding proteinChain: A: PDB Molecule: maltose/maltodextrin transport permease homologue;PDBTitle: maltose/maltodextrin-binding domain male from bdellovibrio2 bacteriovorus bound to maltotriose
68 c4pqkA_
not modelled
100.0
16
PDB header: dna binding proteinChain: A: PDB Molecule: maltose abc transporter periplasmic protein, truncatedPDBTitle: c-terminal domain of dna binding protein
69 c3mp6A_
not modelled
100.0
16
PDB header: histone binding proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,linker,saga-associatedPDBTitle: complex structure of sgf29 and dimethylated h3k4
70 c5y2gA_
not modelled
100.0
16
PDB header: toxinChain: A: PDB Molecule: maltose-binding periplasmic protein,protein b;PDBTitle: structure of mbp tagged gbs camp
71 c3oaiB_
not modelled
100.0
16
PDB header: membrane protein, cell adhesionChain: B: PDB Molecule: maltose-binding periplasmic protein, myelin protein p0;PDBTitle: crystal structure of the extra-cellular domain of human myelin protein2 zero
72 c4bl9D_
not modelled
100.0
16
PDB header: signaling proteinChain: D: PDB Molecule: maltose-binding periplasmic protein, suppressor of fusedPDBTitle: crystal structure of full-length human suppressor of fused (sufu)2 mutant lacking a regulatory subdomain (crystal form i)
73 c4logA_
not modelled
100.0
16
PDB header: transcriptionChain: A: PDB Molecule: maltose abc transporter periplasmic protein and nr2e3PDBTitle: the crystal structure of the orphan nuclear receptor pnr ligand2 binding domain fused with mbp
74 c2vgqA_
not modelled
100.0
16
PDB header: immune system/transportChain: A: PDB Molecule: sugar abc transporter substrate-binding protein,PDBTitle: crystal structure of human ips-1 card
75 c3woaA_
not modelled
100.0
16
PDB header: dna binding protein, sugar binding proteChain: A: PDB Molecule: repressor protein ci, maltose-binding periplasmic protein;PDBTitle: crystal structure of lambda repressor (1-45) fused with maltose-2 binding protein
76 c4rwgC_
not modelled
100.0
16
PDB header: membrane protein/hormoneChain: C: PDB Molecule: maltose-binding periplasmic protein, receptor activity-PDBTitle: crystal structure of the clr:ramp1 extracellular domain heterodimer2 with bound high affinity cgrp analog
77 c6apxA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: maltose-binding periplasmic protein,dual specificityPDBTitle: crystal structure of human dual specificity phosphatase 1 catalytic2 domain (c258s) as a maltose binding protein fusion in complex with3 the monobody ysx1
78 c5ttdA_
not modelled
100.0
15
PDB header: cell adhesionChain: A: PDB Molecule: maltose-binding periplasmic protein,pilin isopeptidePDBTitle: minor pilin fctb from s. pyogenes with engineered intramolecular2 isopeptide bond
79 c4my2A_
not modelled
100.0
16
PDB header: signaling proteinChain: A: PDB Molecule: maltose-binding periplasmic protein, norrin fusion protein;PDBTitle: crystal structure of norrin in fusion with maltose binding protein
80 c4exkA_
not modelled
100.0
15
PDB header: transport proteinChain: A: PDB Molecule: maltose-binding periplasmic protein, uncharacterizedPDBTitle: a chimera protein containing mbp fused to the c-terminal domain of the2 uncharacterized protein stm14_2015 from salmonella enterica
81 c6h0hB_
not modelled
100.0
15
PDB header: sugar binding proteinChain: B: PDB Molecule: probable solute binding protein of abc transporter systemPDBTitle: the abc transporter associated binding protein from b. animalis subsp.2 lactis bl-04 in complex with beta-1,6-galactobiose
82 c5hz7A_
not modelled
100.0
17
PDB header: dna binding proteinChain: A: PDB Molecule: comp;PDBTitle: high-resolution crystal structure of the minor dna-binding pilin comp2 from neisseria meningitidis in fusion with mbp
83 c2zykA_
not modelled
100.0
13
PDB header: sugar binding proteinChain: A: PDB Molecule: solute-binding protein;PDBTitle: crystal structure of cyclo/maltodextrin-binding protein2 complexed with gamma-cyclodextrin
84 c5ixpA_
not modelled
100.0
11
PDB header: transport proteinChain: A: PDB Molecule: extracellular solute-binding protein family 1;PDBTitle: crystal structure of extracellular solute-binding protein family 1
85 c2nvuB_
not modelled
100.0
16
PDB header: protein turnover, ligaseChain: B: PDB Molecule: maltose binding protein/nedd8-activating enzymePDBTitle: structure of appbp1-uba3~nedd8-nedd8-mgatp-ubc12(c111a), a2 trapped ubiquitin-like protein activation complex
86 c3csgA_
not modelled
100.0
17
PDB header: de novo protein, sugar binding proteinChain: A: PDB Molecule: maltose-binding protein monobody ys1 fusion;PDBTitle: crystal structure of monobody ys1(mbp-74)/maltose binding protein2 fusion complex
87 c5k2yB_
not modelled
100.0
18
PDB header: transport proteinChain: B: PDB Molecule: probable periplasmic sugar-binding lipoprotein uspc;PDBTitle: crystal structure of m. tuberculosis uspc (monoclinic crystal form)
88 c1y4cA_
not modelled
100.0
16
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
89 c4wviA_
not modelled
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: maltose-binding periplasmic protein,signal peptidase ib;PDBTitle: crystal structure of the type-i signal peptidase from staphylococcus2 aureus (spsb) in complex with a substrate peptide (pep2).
90 c5yevB_
not modelled
100.0
16
PDB header: apoptosisChain: B: PDB Molecule: tnfrsf25 death domain;PDBTitle: murine dr3 death domain
91 c4ozqA_
not modelled
100.0
16
PDB header: motor proteinChain: A: PDB Molecule: chimera of maltose-binding periplasmic protein and kinesinPDBTitle: crystal structure of the mouse kif14 motor domain
92 c3zkkA_
not modelled
100.0
15
PDB header: transport proteinChain: A: PDB Molecule: xos binding protein;PDBTitle: structure of the xylo-oligosaccharide specific solute binding protein2 from bifidobacterium animalis subsp. lactis bl-04 in complex with3 xylotetraose
93 c3vxbA_
not modelled
100.0
14
PDB header: sugar binding proteinChain: A: PDB Molecule: putative sugar-binding lipoprotein;PDBTitle: crystal structure of bxle from streptomyces thermoviolaceus opc-520
94 c4r0yA_
not modelled
100.0
15
PDB header: protein bindingChain: A: PDB Molecule: maltose-binding periplasmic protein, disks large-associatedPDBTitle: structure of maltose-binding protein fusion with the c-terminal gh12 domain of guanylate kinase-associated protein from rattus norvegicus
95 c4ifpC_
not modelled
100.0
17
PDB header: immune systemChain: C: PDB Molecule: maltose-binding periplasmic protein,nacht, lrr and pydPDBTitle: x-ray crystal structure of human nlrp1 card domain
96 c4r2bB_
not modelled
100.0
14
PDB header: transport proteinChain: B: PDB Molecule: extracellular solute-binding protein family 1;PDBTitle: crystal structure of sugar transporter oant_3817 from ochrobactrum2 anthropi, target efi-510528, with bound glucose
97 c5tu0A_
not modelled
100.0
15
PDB header: transport proteinChain: A: PDB Molecule: lmo2125 protein;PDBTitle: 1.9 angstrom resolution crystal structure of maltose-binding2 periplasmic protein male from listeria monocytogenes in complex with3 maltose
98 c4hs7A_
not modelled
100.0
14
PDB header: solute-binding proteinChain: A: PDB Molecule: bacterial extracellular solute-binding protein, putative;PDBTitle: 2.6 angstrom structure of the extracellular solute-binding protein2 from staphylococcus aureus in complex with peg.
99 c6eg3A_
not modelled
100.0
16
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: maltose/maltodextrin-binding periplasmic protein,probablePDBTitle: crystal structure of human brm in complex with compound 15
100 c2gh9A_
not modelled
100.0
18
PDB header: sugar binding proteinChain: A: PDB Molecule: maltose/maltodextrin-binding protein;PDBTitle: thermus thermophilus maltotriose binding protein bound with2 maltotriose
101 c5dvjA_
not modelled
100.0
14
PDB header: transport proteinChain: A: PDB Molecule: binding protein component of abc sugar transporter;PDBTitle: crystal structure of galactose complexed periplasmic glucose binding2 protein (ppgbp) from p. putida csv86
102 c3ehuA_
not modelled
100.0
15
PDB header: membrane proteinChain: A: PDB Molecule: fusion protein of crfr1 extracellular domain and mbp;PDBTitle: crystal structure of the extracellular domain of human corticotropin2 releasing factor receptor type 1 (crfr1) in complex with crf
103 c3oo6A_
not modelled
100.0
15
PDB header: sugar binding proteinChain: A: PDB Molecule: abc transporter binding protein acbh;PDBTitle: crystal structures and biochemical characterization of the bacterial2 solute receptor acbh reveal an unprecedented exclusive substrate3 preference for b-d-galactopyranose
104 d1elja_
not modelled
100.0
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
105 c4rk9B_
not modelled
100.0
12
PDB header: transport proteinChain: B: PDB Molecule: carbohydrate abc transporter substrate-binding proteinPDBTitle: crystal structure of sugar transporter bl01359 from bacillus2 licheniformis, target efi-510856, in complex with stachyose
106 c5az6A_
not modelled
100.0
15
PDB header: sugar binding protein, peptide binding pChain: A: PDB Molecule: maltose-binding periplasmic protein,mitochondrial importPDBTitle: crystal structure of mbp-tom20 fusion protein with a 2-residue spacer2 in the connector helix
107 c4rk2B_
not modelled
100.0
18
PDB header: transport proteinChain: B: PDB Molecule: putative sugar abc transporter, substrate-binding protein;PDBTitle: crystal structure of sugar transporter rhe_pf00321 from rhizobium2 etli, target efi-510806, an open conformation
108 c3a3cA_
not modelled
100.0
15
PDB header: protein transportChain: A: PDB Molecule: maltose-binding periplasmic protein, linker, mitochondrialPDBTitle: crystal structure of tim40/mia40 fusing mbp, c296s and c298s mutant
109 c2b3fD_
not modelled
100.0
13
PDB header: sugar binding proteinChain: D: PDB Molecule: glucose-binding protein;PDBTitle: thermus thermophilus glucose/galactose binding protein bound with2 galactose
110 c2w7yA_
not modelled
100.0
16
PDB header: sugar-binding proteinChain: A: PDB Molecule: probable sugar abc transporter, sugar-bindingPDBTitle: structure of a streptococcus pneumoniae solute-binding2 protein in complex with the blood group a-trisaccharide.
111 c5c7rA_
not modelled
100.0
16
PDB header: antifreeze proteinChain: A: PDB Molecule: fusion protein of maltose-binding periplasmic protein andPDBTitle: revealing surface waters on an antifreeze protein by fusion protein2 crystallography
112 c1mg1A_
not modelled
100.0
16
PDB header: viral proteinChain: A: PDB Molecule: protein (htlv-1 gp21 ectodomain/maltose-binding proteinPDBTitle: htlv-1 gp21 ectodomain/maltose-binding protein chimera
113 c4o2xA_
not modelled
100.0
17
PDB header: transport proteinChain: A: PDB Molecule: maltose-binding periplasmic protein, atp-dependent clpPDBTitle: structure of a malarial protein
114 c2fncA_
not modelled
100.0
17
PDB header: sugar binding proteinChain: A: PDB Molecule: maltose abc transporter, periplasmic maltose-bindingPDBTitle: thermotoga maritima maltotriose binding protein bound with2 maltotriose.
115 c2xd3A_
not modelled
100.0
15
PDB header: sugar binding proteinChain: A: PDB Molecule: maltose/maltodextrin-binding protein;PDBTitle: the crystal structure of malx from streptococcus pneumoniae2 in complex with maltopentaose.
116 c5b3zB_
not modelled
100.0
14
PDB header: isomerase,sugar binding proteinChain: B: PDB Molecule: peptidyl-prolyl cis-trans isomerase nima-interacting 1,PDBTitle: crystal structure of hpin1 ww domain (5-39) fused with maltose-binding2 protein
117 c4ua8A_
not modelled
100.0
14
PDB header: transport proteinChain: A: PDB Molecule: carbohydrate abc transporter substrate-binding protein,PDBTitle: eur_01830 (maltotriose-binding protein) complexed with maltotriose
118 c3i3vC_
not modelled
100.0
15
PDB header: transport proteinChain: C: PDB Molecule: probable secreted solute-binding lipoprotein;PDBTitle: crystal structure of probable secreted solute-binding2 lipoprotein from streptomyces coelicolor
119 c1mh3A_
not modelled
100.0
16
PDB header: sugar binding, dna binding proteinChain: A: PDB Molecule: maltose binding-a1 homeodomain protein chimera;PDBTitle: maltose binding-a1 homeodomain protein chimera, crystal form i
120 c1ursA_
not modelled
100.0
18
PDB header: maltose-binding proteinChain: A: PDB Molecule: maltose-binding protein;PDBTitle: x-ray structures of the maltose-maltodextrin binding2 protein of the thermoacidophilic bacterium alicyclobacillus3 acidocaldarius