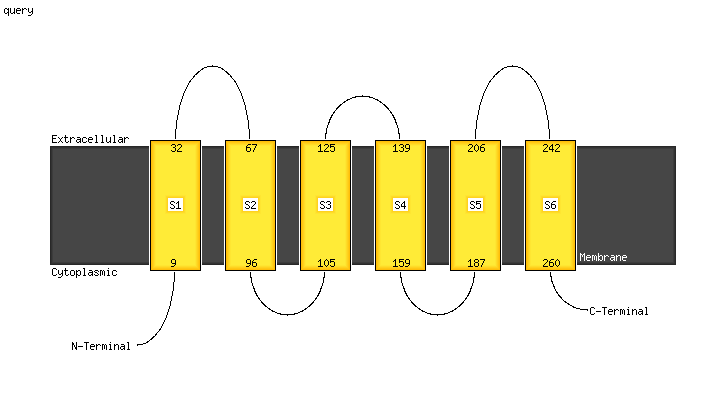
1 c4tqvJ_
100.0
25
PDB header: transport proteinChain: J: PDB Molecule: algm2;PDBTitle: crystal structure of a bacterial abc transporter involved in the2 import of the acidic polysaccharide alginate
2 d2r6gg1
100.0
29
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like
3 c3fh6F_
100.0
15
PDB header: transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
4 c4tqvI_
100.0
17
PDB header: transport proteinChain: I: PDB Molecule: algm1;PDBTitle: crystal structure of a bacterial abc transporter involved in the2 import of the acidic polysaccharide alginate
5 c2r6gF_
100.0
18
PDB header: hydrolase/transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: the crystal structure of the e. coli maltose transporter
6 d2onkc1
100.0
17
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like
7 c2onkC_
100.0
17
PDB header: membrane proteinChain: C: PDB Molecule: molybdate/tungstate abc transporter, permeasePDBTitle: abc transporter modbc in complex with its binding protein2 moda
8 d2r6gf2
100.0
19
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like
9 d3d31c1
100.0
20
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like
10 c3d31D_
100.0
20
PDB header: transport proteinChain: D: PDB Molecule: sulfate/molybdate abc transporter, permease protein;PDBTitle: modbc from methanosarcina acetivorans
11 c4ymuC_
100.0
13
PDB header: protein binding/transport proteinChain: C: PDB Molecule: abc-type amino acid transport system, permease component;PDBTitle: crystal structure of an amino acid abc transporter complex with2 arginines and atps
12 d3dhwa1
99.9
21
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like
13 c5kbuA_
76.9
14
PDB header: transport proteinChain: A: PDB Molecule: glutamate receptor 2,voltage-dependent calcium channelPDBTitle: cryo-em structure of glua2-2xstz complex at 7.8 angstrom resolution
14 d2r6gf1
70.4
11
Fold: MalF N-terminal region-likeSuperfamily: MalF N-terminal region-likeFamily: MalF N-terminal region-like
15 c2m8gX_
46.7
19
PDB header: transcriptionChain: X: PDB Molecule: transcriptional regulator;PDBTitle: structure, function, and tethering of dna-binding domains in 542 transcriptional activators
16 c1umqA_
43.7
13
PDB header: dna-binding proteinChain: A: PDB Molecule: photosynthetic apparatus regulatory protein;PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
17 d1umqa_
43.7
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like
18 c4l5eA_
43.5
13
PDB header: protein bindingChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of a. aeolicus ntrc1 dna binding domain
19 d1ntca_
41.3
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like
20 d1fipa_
31.4
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like
21 c3e7lD_
not modelled
26.5
10
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
22 d1etob_
not modelled
26.1
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like
23 d1etxa_
not modelled
25.0
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like
24 c2hx6A_
not modelled
24.6
23
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease;PDBTitle: solution structure analysis of the phage t42 endoribonuclease regb
25 c2jwaA_
not modelled
22.7
21
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
26 c2ks1A_
not modelled
22.7
21
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
27 d1g2ha_
not modelled
16.1
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like
28 c1g2hA_
not modelled
16.1
10
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein tyrr homolog;PDBTitle: solution structure of the dna-binding domain of the tyrr2 protein of haemophilus influenzae
29 c5m7nA_
not modelled
14.3
13
PDB header: signaling proteinChain: A: PDB Molecule: nitrogen assimilation regulatory protein;PDBTitle: crystal structure of ntrx from brucella abortus in complex with atp2 processed with the crystaldirect automated mounting and cryo-cooling3 technology
30 c5ir6A_
not modelled
12.3
11
PDB header: oxidoreductaseChain: A: PDB Molecule: bd-type quinol oxidase subunit i;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
31 c1ojlD_
not modelled
11.4
23
PDB header: response regulatorChain: D: PDB Molecule: transcriptional regulatory protein zrar;PDBTitle: crystal structure of a sigma54-activator suggests the mechanism for2 the conformational switch necessary for sigma54 binding
32 c1bctA_
not modelled
9.8
21
PDB header: photoreceptorChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: three-dimensional structure of proteolytic fragment 163-2312 of bacterioopsin determined from nuclear magnetic3 resonance data in solution
33 c4huqS_
not modelled
9.6
11
PDB header: hydrolaseChain: S: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a transporter
34 c3s1bA_
not modelled
8.6
24
PDB header: signaling proteinChain: A: PDB Molecule: mini-z;PDBTitle: the development of peptide-based tools for the analysis of2 angiogenesis
35 c2cw1A_
not modelled
8.6
27
PDB header: de novo proteinChain: A: PDB Molecule: sn4m;PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
36 c2n2aA_
not modelled
7.7
15
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: spatial structure of her2/erbb2 dimeric transmembrane domain in the2 presence of cytoplasmic juxtamembrane domains
37 c4j2nA_
not modelled
7.1
15
PDB header: viral proteinChain: A: PDB Molecule: gp37;PDBTitle: crystal structure of mycobacteriophage pukovnik xis
38 c4j2nB_
not modelled
7.1
15
PDB header: viral proteinChain: B: PDB Molecule: gp37;PDBTitle: crystal structure of mycobacteriophage pukovnik xis
39 c2momB_
not modelled
6.9
19
PDB header: membrane proteinChain: B: PDB Molecule: lysosome-associated membrane glycoprotein 2;PDBTitle: structural insights of tm domain of lamp-2a in dpc micelles
40 c2momC_
not modelled
6.9
19
PDB header: membrane proteinChain: C: PDB Molecule: lysosome-associated membrane glycoprotein 2;PDBTitle: structural insights of tm domain of lamp-2a in dpc micelles
41 c1pyuD_
not modelled
6.9
15
PDB header: lyaseChain: D: PDB Molecule: aspartate 1-decarboxylase alfa chain;PDBTitle: processed aspartate decarboxylase mutant with ser25 mutated to cys
42 c4qiwT_
not modelled
6.9
50
PDB header: transcriptionChain: T: PDB Molecule: dna-directed rna polymerase subunit k;PDBTitle: crystal structure of euryarchaeal rna polymerase from thermococcus2 kodakarensis
43 c1twcF_
not modelled
6.6
43
PDB header: transcriptionChain: F: PDB Molecule: dna-directed rna polymerases i, ii, and iii 23PDBTitle: rna polymerase ii complexed with gtp
44 d1twff_
not modelled
6.5
43
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RPB6
45 c2pmzW_
not modelled
6.3
50
PDB header: translation, transferaseChain: W: PDB Molecule: dna-directed rna polymerase subunit k;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
46 d1cf7a_
not modelled
5.9
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp
47 c4xtnJ_
not modelled
5.5
7
PDB header: membrane proteinChain: J: PDB Molecule: sodium pumping rhodopsin;PDBTitle: crystal structure of the light-driven sodium pump kr2 in the2 pentameric red form, ph 4.9
48 c6eyuA_
not modelled
5.4
14
PDB header: membrane proteinChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: crystal structure of the inward h(+) pump xenorhodopsin
49 d2ns0a1
not modelled
5.3
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like
50 d1qkla_
not modelled
5.3
43
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RPB6